RGSB-UNet: Hybrid Deep Learning Framework for Tumour Segmentation in Digital Pathology Images
- PMID: 37627842
- PMCID: PMC10452008
- DOI: 10.3390/bioengineering10080957
RGSB-UNet: Hybrid Deep Learning Framework for Tumour Segmentation in Digital Pathology Images
Abstract
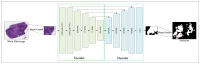
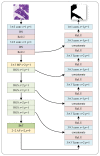
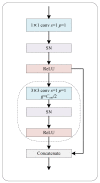
Colorectal cancer (CRC) is a prevalent gastrointestinal tumour with high incidence and mortality rates. Early screening for CRC can improve cure rates and reduce mortality. Recently, deep convolution neural network (CNN)-based pathological image diagnosis has been intensively studied to meet the challenge of time-consuming and labour-intense manual analysis of high-resolution whole slide images (WSIs). Despite the achievements made, deep CNN-based methods still suffer from some limitations, and the fundamental problem is that they cannot capture global features. To address this issue, we propose a hybrid deep learning framework (RGSB-UNet) for automatic tumour segmentation in WSIs. The framework adopts a UNet architecture that consists of the newly-designed residual ghost block with switchable normalization (RGS) and the bottleneck transformer (BoT) for downsampling to extract refined features, and the transposed convolution and 1 × 1 convolution with ReLU for upsampling to restore the feature map resolution to that of the original image. The proposed framework combines the advantages of the spatial-local correlation of CNNs and the long-distance feature dependencies of BoT, ensuring its capacity of extracting more refined features and robustness to varying batch sizes. Additionally, we consider a class-wise dice loss (CDL) function to train the segmentation network. The proposed network achieves state-of-the-art segmentation performance under small batch sizes. Experimental results on DigestPath2019 and GlaS datasets demonstrate that our proposed model produces superior evaluation scores and state-of-the-art segmentation results.
Keywords: Residual-Ghost-SN; bottleneck transformer; hybrid deep learning framework; tumour segmentation; whole slide image.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Song W., Fu C., Zheng Y., Cao L., Tie M. A practical medical image cryptosystem with parallel acceleration. J. Ambient. Intell. Humaniz. Comput. 2022;14:9853–9867. doi: 10.1007/s12652-021-03643-6. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

