Split Hand-Foot and Deafness in a Patient with 7q21.13-q21.3 Deletion Not Including the DLX5/6 Genes
- PMID: 37628577
- PMCID: PMC10454356
- DOI: 10.3390/genes14081526
Split Hand-Foot and Deafness in a Patient with 7q21.13-q21.3 Deletion Not Including the DLX5/6 Genes
Abstract
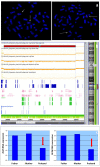
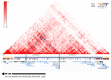
Split Hand-Foot Malformation (SHFM) is a congenital limb defect characterized by a median cleft of the hands and/or feet due to the absence/hypoplasia of the central rays. It may occur as part of a syndromic condition or as an isolated malformation. The most common of the six genetic loci identified for this condition is correlated to SHFM1 and maps in the 7q21q22 region. SHFM1 is characterized by autosomal dominant transmission, incomplete penetrance and variable expressivity. Associated features often include hearing loss, intellectual disability/developmental delay and craniofacial abnormalities. Disruption of the DLX5/DLX6 genes, mapping within the SHFM1 locus, is now known to be responsible for the phenotype. Through SNP array, we analyzed a patient affected by SHFM1 associated with deafness and an abnormality of the inner ear (incomplete partition type I); we identified a deletion in 7q21, not involving the DLX5/6 genes, but including exons 15 and 17 of DYNC1I1, known to act as exonic enhancers (eExons) of the DLX5/6 genes. We further demonstrated the role of DYNC1I1 eExons in regulating DLX5/6 expression by means of showing a reduced expression of the DLX5/6 genes through RT-PCR in a patient-derived lymphoblastoid cell line. Furthermore, our data and a review of published cases do not support the hypothesis that DLX5/6 are imprinted in humans. This work is an example of how the disruption of regulatory elements can be responsible for congenital malformations.
Keywords: DLX5; DLX6; DYNC1I1; SHFM; TADs; ectrodactyly; imprinting; regulatory elements.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Bernardini L., Palka C., Ceccarini C., Capalbo A., Bottillo I., Mingarelli R., Novelli A., Dallapiccola B. Complex rearrangement of chromosomes 7q21.13-q22.1 confirms the ectrodactyly-deafness locus and suggests new candidate genes. Am. J. Med. Genet. Part A. 2007;146A:238–244. doi: 10.1002/ajmg.a.32093. - DOI - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical

