Amplicon-Based Microbiome Profiling: From Second- to Third-Generation Sequencing for Higher Taxonomic Resolution
- PMID: 37628619
- PMCID: PMC10454624
- DOI: 10.3390/genes14081567
Amplicon-Based Microbiome Profiling: From Second- to Third-Generation Sequencing for Higher Taxonomic Resolution
Abstract
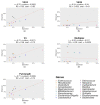
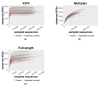
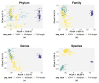
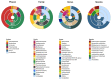
The 16S rRNA amplicon-based sequencing approach represents the most common and cost-effective strategy with great potential for microbiome profiling. The use of second-generation sequencing (NGS) technologies has led to protocols based on the amplification of one or a few hypervariable regions, impacting the outcome of the analysis. Nowadays, comparative studies are necessary to assess different amplicon-based approaches, including the full-locus sequencing currently feasible thanks to third-generation sequencing (TGS) technologies. This study compared three different methods to achieve the deepest microbiome taxonomic characterization: (a) the single-region approach, (b) the multiplex approach, covering several regions of the target gene/region, both based on NGS short reads, and (c) the full-length approach, which analyzes the whole length of the target gene thanks to TGS long reads. Analyses carried out on benchmark microbiome samples, with a known taxonomic composition, highlighted a different classification performance, strongly associated with the type of hypervariable regions and the coverage of the target gene. Indeed, the full-length approach showed the greatest discriminating power, up to species level, also on complex real samples. This study supports the transition from NGS to TGS for the study of the microbiome, even if experimental and bioinformatic improvements are still necessary.
Keywords: 16S rRNA amplicon-based sequencing; metagenomics; microbiome; mock analysis; next-generation sequencing; third-generation sequencing.
Conflict of interest statement
N.T. is employed as a scientist by Postbiotica S.r.l. M.R. is one of the founders of Postbiotica S.r.l. M.R. is chief scientific officer of Postbiotica. The authors declare no conflict of interest.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

