Examination of Factors Affecting Site-Directed RNA Editing by the MS2-ADAR1 Deaminase System
- PMID: 37628635
- PMCID: PMC10454654
- DOI: 10.3390/genes14081584
Examination of Factors Affecting Site-Directed RNA Editing by the MS2-ADAR1 Deaminase System
Abstract
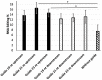
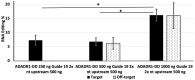
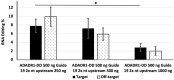
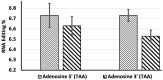
Adenosine deaminases acting on RNA (ADARs) have double-stranded RNA binding domains and a deaminase domain (DD). We used the MS2 system and specific guide RNAs to direct ADAR1-DD to target adenosines in the mRNA encoding-enhanced green fluorescence protein. Using this system in transfected HEK-293 cells, we evaluated the effects of changing the length and position of the guide RNA on the efficiency of conversion of amber (TAG) and ochre (TAA) stop codons to tryptophan (TGG) in the target. Guide RNAs of 19, 21 and 23 nt were positioned upstream and downstream of the MS2-RNA, providing a total of six guide RNAs. The upstream guide RNAs were more functionally effective than the downstream guide RNAs, with the following hierarchy of efficiency: 21 nt > 23 nt > 19 nt. The highest editing efficiency was 16.6%. Off-target editing was not detected in the guide RNA complementary region but was detected 50 nt downstream of the target. The editing efficiency was proportional to the amount of transfected deaminase but inversely proportional to the amount of the transfected guide RNA. Our results suggest that specific RNA editing requires precise optimization of the ratio of enzyme, guide RNA, and target RNA.
Keywords: RNA editing; efficiency; gene therapy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

