Changes in m6A in Steatotic Liver Disease
- PMID: 37628704
- PMCID: PMC10454815
- DOI: 10.3390/genes14081653
Changes in m6A in Steatotic Liver Disease
Abstract
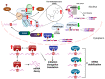
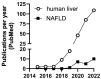
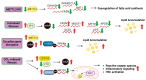
Fatty liver disease is one of the major causes of morbidity and mortality worldwide. Fatty liver includes non-alcoholic fatty liver disease (NAFLD) and non-alcoholic steatohepatitis (NASH), now replaced by a consensus group as metabolic dysfunction-associated steatotic liver disease (MASLD). While excess nutrition and obesity are major contributors to fatty liver, the underlying mechanisms remain largely unknown and therapeutic interventions are limited. Reversible chemical modifications in RNA are newly recognized critical regulators controlling post-transcriptional gene expression. Among these modifications, N6-methyladenosine (m6A) is the most abundant and regulates transcript abundance in fatty liver disease. Modulation of m6A by readers, writers, and erasers (RWE) impacts mRNA processing, translation, nuclear export, localization, and degradation. While many studies focus on m6A RWE expression in human liver pathologies, limitations of technology and bioinformatic methods to detect m6A present challenges in understanding the epitranscriptomic mechanisms driving fatty liver disease progression. In this review, we summarize the RWE of m6A and current methods of detecting m6A in specific genes associated with fatty liver disease.
Keywords: NAFLD; NASH; RNA modifications; epitranscriptome; fatty liver; m6A.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Rinella M.E., Lazarus J.V., Ratziu V., Francque S.M., Sanyal A.J., Kanwal F., Romero D., Abdelmalek M.F., Anstee Q.M., Arab J.P., et al. A multi-society Delphi consensus statement on new fatty liver disease nomenclature. J. Hepatol. 2023 online ahead of print . - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

