HmbC, a Protein of the HMG Family, Participates in the Regulation of Carotenoid Biosynthesis in Fusarium fujikuroi
- PMID: 37628712
- PMCID: PMC10454146
- DOI: 10.3390/genes14081661
HmbC, a Protein of the HMG Family, Participates in the Regulation of Carotenoid Biosynthesis in Fusarium fujikuroi
Abstract
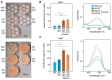
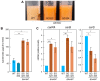
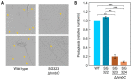
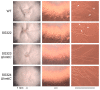
In the fungus Fusarium fujikuroi, carotenoid production is up-regulated by light and down-regulated by the CarS RING finger protein, which modulates the mRNA levels of carotenoid pathway genes (car genes). To identify new potential regulators of car genes, we used a biotin-mediated pull-down procedure to detect proteins capable of binding to their promoters. We focused our attention on one of the proteins found in the screening, belonging to the High-Mobility Group (HMG) family that was named HmbC. The deletion of the hmbC gene resulted in increased carotenoid production due to higher mRNA levels of car biosynthetic genes. In addition, the deletion resulted in reduced carS mRNA levels, which could also explain the partial deregulation of the carotenoid pathway. The mutants exhibited other phenotypic traits, such as alterations in development under certain stress conditions, or reduced sensitivity to cell wall degrading enzymes, revealed by less efficient protoplast formation, indicating that HmbC is also involved in other cellular processes. In conclusion, we identified a protein of the HMG family that participates in the regulation of carotenoid biosynthesis. This is probably achieved through an epigenetic mechanism related to chromatin structure, as is frequent in this class of proteins.
Keywords: HmbC protein; carS gene; carotenoids; high-mobility group; protoplast formation; pull-down assay.
Conflict of interest statement
S.N. is employed by the Symrise AG Chemicals company, Holzminden, Germany. The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures

Similar articles
-
Comparative transcriptomic analysis unveils interactions between the regulatory CarS protein and light response in Fusarium.BMC Genomics. 2019 Jan 21;20(1):67. doi: 10.1186/s12864-019-5430-x. BMC Genomics. 2019. PMID: 30665350 Free PMC article.
-
Genetic basis of carotenoid overproduction in Fusarium oxysporum.Fungal Genet Biol. 2012 Sep;49(9):684-96. doi: 10.1016/j.fgb.2012.06.007. Epub 2012 Jun 26. Fungal Genet Biol. 2012. PMID: 22750191
-
Functional analysis of the carS gene of Fusarium fujikuroi.Mol Genet Genomics. 2013 Apr;288(3-4):157-73. doi: 10.1007/s00438-013-0739-7. Epub 2013 Mar 30. Mol Genet Genomics. 2013. PMID: 23543145
-
Carotenoid Biosynthesis in Fusarium.J Fungi (Basel). 2017 Jul 7;3(3):39. doi: 10.3390/jof3030039. J Fungi (Basel). 2017. PMID: 29371556 Free PMC article. Review.
-
Carotenoids, versatile components of oxygenic photosynthesis.Prog Lipid Res. 2013 Oct;52(4):539-61. doi: 10.1016/j.plipres.2013.07.001. Epub 2013 Jul 26. Prog Lipid Res. 2013. PMID: 23896007 Review.
Cited by
-
The Non-Histone Protein FgNhp6 Is Involved in the Regulation of the Development, DON Biosynthesis, and Virulence of Fusarium graminearum.Pathogens. 2024 Jul 16;13(7):592. doi: 10.3390/pathogens13070592. Pathogens. 2024. PMID: 39057819 Free PMC article.
References
-
- Mallik R., Kundu A., Chaudhuri S. High Mobility Group Proteins: The Multifaceted Regulators of Chromatin Dynamics. Nucleus. 2018;61:213–226. doi: 10.1007/s13237-018-0257-4. - DOI
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources

