Shedding Light on the Photophysics and Photochemistry of I-Motifs Using Quantum Mechanical Calculations
- PMID: 37628797
- PMCID: PMC10454157
- DOI: 10.3390/ijms241612614
Shedding Light on the Photophysics and Photochemistry of I-Motifs Using Quantum Mechanical Calculations
Abstract
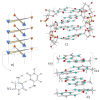
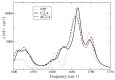
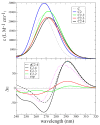
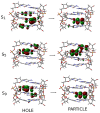
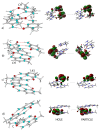
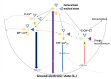
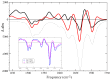
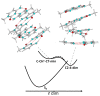
I-motifs are non-canonical DNA structures formed by intercalated hemiprotonated (CH·C)+ pairs, i.e., formed by a cytosine (C) and a protonated cytosine (CH+), which are currently drawing great attention due to their biological relevance and promising nanotechnological properties. It is important to characterize the processes occurring in I-motifs following irradiation by UV light because they can lead to harmful consequences for genetic code and because optical spectroscopies are the most-used tools to characterize I-motifs. By using time-dependent DFT calculations, we here provide the first comprehensive picture of the photoactivated behavior of the (CH·C)+ core of I-motifs, from absorption to emission, while also considering the possible photochemical reactions. We reproduce and assign their spectral signatures, i.e., infrared, absorption, fluorescence and circular dichroism spectra, disentangling the underlying chemical-physical effects. We show that the main photophysical paths involve C and CH+ bases on adjacent steps and, using this basis, interpret the available time-resolved spectra. We propose that a photodimerization reaction can occur on an excited state with strong C→CH+ charge transfer character and examine some of the possible photoproducts. Based on the results reported, some future perspectives for the study of I-motifs are discussed.
Keywords: TD-DFT; non-canonical DNA structure; protonated cytosine.
Conflict of interest statement
The author declares no conflict of interest.
Figures
Similar articles
-
The photophysics of protonated cytidine and hemiprotonated cytidine base pair: A computational study.Photochem Photobiol. 2024 Mar-Apr;100(2):314-322. doi: 10.1111/php.13832. Epub 2023 Jul 6. Photochem Photobiol. 2024. PMID: 37409732
-
Nucleic Acids as a Playground for the Computational Study of the Photophysics and Photochemistry of Multichromophore Assemblies.Acc Chem Res. 2022 Aug 2;55(15):2077-2087. doi: 10.1021/acs.accounts.2c00256. Epub 2022 Jul 14. Acc Chem Res. 2022. PMID: 35833758
-
The photoactivated dynamics of dGpdC and dCpdG sequences in DNA: a comprehensive quantum mechanical study.Chem Sci. 2024 May 16;15(25):9676-9693. doi: 10.1039/d4sc00910j. eCollection 2024 Jun 26. Chem Sci. 2024. PMID: 38939156 Free PMC article.
-
Quantum Mechanical Studies on the Photophysics and the Photochemistry of Nucleic Acids and Nucleobases.Chem Rev. 2016 Mar 23;116(6):3540-93. doi: 10.1021/acs.chemrev.5b00444. Epub 2016 Mar 1. Chem Rev. 2016. PMID: 26928320 Review.
-
Studying the excited electronic states of guanine rich DNA quadruplexes by quantum mechanical methods: main achievements and perspectives.Photochem Photobiol Sci. 2020 Apr 15;19(4):436-444. doi: 10.1039/d0pp00065e. Photochem Photobiol Sci. 2020. PMID: 32255446 Review.
Cited by
-
Stabilization of G-Quadruplex Structures of the SARS-CoV-2 Genome by TMPyP4, BRACO19, and PhenDC3.Int J Mol Sci. 2024 Feb 20;25(5):2482. doi: 10.3390/ijms25052482. Int J Mol Sci. 2024. PMID: 38473730 Free PMC article.
References
-
- Mergny J.L., Lacroix L., Han X., Leroy J.L., Helene C. Intramolecular Folding of Pyrimidine Oligodeoxynucleotides into an i-DNA Motif. J. Am. Chem. Soc. 1995;117:8887–8898. doi: 10.1021/ja00140a001. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

