Inhibition of USP2 Enhances TRAIL-Mediated Cancer Cell Death through Downregulation of Survivin
- PMID: 37628997
- PMCID: PMC10454696
- DOI: 10.3390/ijms241612816
Inhibition of USP2 Enhances TRAIL-Mediated Cancer Cell Death through Downregulation of Survivin
Abstract
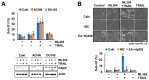
Ubiquitin-specific protease 2 (USP2) is a deubiquitinase belonging to the USPs subfamily. USP2 has been known to display various biological effects including tumorigenesis and inflammation. Therefore, we aimed to examine the sensitization effect of USP2 in TRAIL-mediated apoptosis. The pharmacological inhibitor (ML364) and siRNA targeting USP2 enhanced TNF-related apoptosis-inducing ligand (TRAIL)-induced cancer cell death, but not normal cells. Mechanistically, USP2 interacted with survivin, and ML364 degraded survivin protein expression by increasing the ubiquitination of survivin. Overexpression of survivin or USP2 significantly prevented apoptosis through cotreatment with ML364 and TRAIL, whereas a knockdown of USP2 increased sensitivity to TRAIL. Taken together, our data suggested that ML364 ubiquitylates and degrades survivin, thereby increasing the reactivity to TRAIL-mediated apoptosis in cancer cells.
Keywords: ML364; TRAIL; USP2; deubiquitinase; survivin.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
Cepharanthine Enhances TRAIL-Mediated Apoptosis Through STAMBPL1-Mediated Downregulation of Survivin Expression in Renal Carcinoma Cells.Int J Mol Sci. 2018 Oct 22;19(10):3280. doi: 10.3390/ijms19103280. Int J Mol Sci. 2018. PMID: 30360403 Free PMC article.
-
Pharmacological USP2 targeting suppresses ovarian cancer growth by potentiating apoptosis and ferroptosis.Arch Biochem Biophys. 2024 Dec;762:110193. doi: 10.1016/j.abb.2024.110193. Epub 2024 Oct 30. Arch Biochem Biophys. 2024. PMID: 39486565
-
Axl Inhibitor R428 Enhances TRAIL-Mediated Apoptosis Through Downregulation of c-FLIP and Survivin Expression in Renal Carcinoma.Int J Mol Sci. 2019 Jul 2;20(13):3253. doi: 10.3390/ijms20133253. Int J Mol Sci. 2019. PMID: 31269715 Free PMC article.
-
Down-regulation of intracellular anti-apoptotic proteins, particularly c-FLIP by therapeutic agents; the novel view to overcome resistance to TRAIL.J Cell Physiol. 2018 Oct;233(10):6470-6485. doi: 10.1002/jcp.26585. Epub 2018 May 9. J Cell Physiol. 2018. PMID: 29741767 Review.
-
Targeting the deubiquitinase USP2 for malignant tumor therapy (Review).Oncol Rep. 2023 Oct;50(4):176. doi: 10.3892/or.2023.8613. Epub 2023 Aug 18. Oncol Rep. 2023. PMID: 37594087 Free PMC article. Review.
Cited by
-
VCP enhances autophagy-related osteosarcoma progression by recruiting USP2 to inhibit ubiquitination and degradation of FASN.Cell Death Dis. 2024 Nov 3;15(11):788. doi: 10.1038/s41419-024-07168-6. Cell Death Dis. 2024. PMID: 39489738 Free PMC article.
-
Lovastatin Targets the USP14-Survivin Axis to Suppress Triple-Negative Breast Cancer via Ubiquitin-Mediated Proteasomal Degradation.Cells. 2025 May 31;14(11):816. doi: 10.3390/cells14110816. Cells. 2025. PMID: 40497992 Free PMC article.
-
Deubiquitinating enzyme USP2 regulates brown adipose tissue thermogenesis via controlling EBF2 stabilization.Mol Metab. 2025 Jun;96:102139. doi: 10.1016/j.molmet.2025.102139. Epub 2025 Apr 4. Mol Metab. 2025. PMID: 40189098 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

