An Outbred Calf Model for Determining Innate Immune Sensing and Evolutionary Trajectories of a Cell Culture-Adapted Bovine Foamy Virus Variant
- PMID: 37632114
- PMCID: PMC10458543
- DOI: 10.3390/v15081772
An Outbred Calf Model for Determining Innate Immune Sensing and Evolutionary Trajectories of a Cell Culture-Adapted Bovine Foamy Virus Variant
Abstract
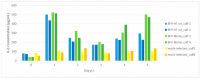
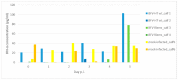
Bovine foamy virus (BFVbta) displays a very high degree of cell-associated replication which is unprecedented even among the other known foamy viruses. Interestingly, recent studies have shown that it can in fact adapt in vitro to high-titer (HT) cell-free transmission due to genetic changes acquired during repeated rounds of cell-free BFVbta passages in immortalized bovine MDBK cells. Molecular clones obtained from the HT BFVbta Riems cell-free variant (HT BFVbta Riems) have been thoroughly characterized in MDBK cell cultures However, during recent years, it has become increasingly clear that the source of the host cells used for virus growth and functional studies of virus replication and virus-cell interactions plays a paramount role. Established cell lines, mostly derived from tumors, but occasionally experimentally immortalized and transformed, frequently display aberrant features relating, for example. to growth, metabolism, and genetics. Even state-of-the-art organoid cultures of primary cells cannot replicate the conditions in an authentic host, especially those concerning cell diversity and the role of innate and adaptive immunity. Therefore, to determine the overall replication characteristics of the cloned wt and HT BFVbta Riems variant, we conducted a small-scale animal pilot study. The replication of the original wt BFVbta Riems isolate, as well as that of its HT variant, were analyzed. Both BFVbta variants established infection in calves, with proviruses in peripheral blood mononuclear cells and induced Gag-specific antibodies. In addition, a related pattern in the host innate immune reaction was detected in the peripheral blood leukocytes of the BFV-infected calves. Surprisingly, an analysis of the Gag sequence two weeks post-inoculation revealed that the HT BFVbta variant showed a very high level of genetic reversion to the wild type (parental BFVbta genotype).
Keywords: bovine foamy virus; calves; cell-free and cell-associated variants; replication in vivo.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
In Vitro Evolution of Bovine Foamy Virus Variants with Enhanced Cell-Free Virus Titers and Transmission.Viruses. 2015 Nov 11;7(11):5855-74. doi: 10.3390/v7112907. Viruses. 2015. PMID: 26569290 Free PMC article.
-
Similar patterns of infection with bovine foamy virus in experimentally inoculated calves and sheep.J Virol. 2013 Mar;87(6):3516-25. doi: 10.1128/JVI.02447-12. Epub 2013 Jan 16. J Virol. 2013. PMID: 23325680 Free PMC article.
-
Complete genome sequences of two novel European clade bovine foamy viruses from Germany and Poland.J Virol. 2012 Oct;86(19):10905-6. doi: 10.1128/JVI.01875-12. J Virol. 2012. PMID: 22966195 Free PMC article.
-
Non-primate foamy viruses.Curr Top Microbiol Immunol. 2003;277:197-211. doi: 10.1007/978-3-642-55701-9_9. Curr Top Microbiol Immunol. 2003. PMID: 12908774 Review.
-
[How the bovine viral diarrhea virus outwits the immune system].Dtsch Tierarztl Wochenschr. 2006 Apr;113(4):124-9. Dtsch Tierarztl Wochenschr. 2006. PMID: 16716045 Review. German.
References
-
- Bao Q., Hotz-Wagenblatt A., Betts M.J., Hipp M., Hugo A., Pougialis G., Lei-Rossmann J., Löchelt M. Shared and cell type-specific adaptation strategies of Gag and Env yield high titer bovine foamy virus variants. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2020;82:104287. doi: 10.1016/j.meegid.2020.104287. - DOI - PubMed
-
- Materniak M., Sieradzki Z., Kuźmak J. Detection of bovine foamy virus in milk and saliva of BFV seropositive cattle. Bull Vet Inst Pulawy. 2010;54:461–465.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

