Assessing the Intense Influenza A(H1N1)pdm09 Epidemic and Vaccine Effectiveness in the Post-COVID Season in the Russian Federation
- PMID: 37632122
- PMCID: PMC10458445
- DOI: 10.3390/v15081780
Assessing the Intense Influenza A(H1N1)pdm09 Epidemic and Vaccine Effectiveness in the Post-COVID Season in the Russian Federation
Abstract
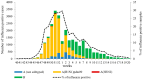
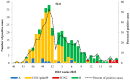
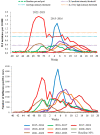
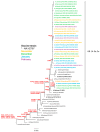
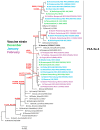
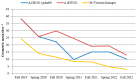
The COVID-19 pandemic had a profound impact on influenza activity worldwide. However, as the pandemic progressed, influenza activity resumed. Here, we describe the influenza epidemic of high intensity of the 2022-2023 season. The epidemic had an early start and peaked in week 51.2022. The extremely high intensity of the epidemic may have been due to a significant decrease in herd immunity. The results of PCR-testing of 220,067 clinical samples revealed that the influenza A(H1N1)pdm09 virus dominated, causing 56.4% of positive cases, while A(H3N2) influenza subtype accounted for only 0.6%, and influenza B of Victoria lineage-for 34.3%. The influenza vaccine was found to be highly effective, with an estimated effectiveness of 92.7% in preventing admission with laboratory-confirmed influenza severe acute respiratory illness (SARI) cases and 54.7% in preventing influenza-like illness/acute respiratory illness (ILI/ARI) cases due to antigenic matching of circulated viruses with influenza vaccine strains for the season. Full genome next-generation sequencing of 1723 influenza A(H1N1)pdm09 viruses showed that all of them fell within clade 6B.1A.5.a2; nine of them possessed H275Y substitution in the NA gene, a genetic marker of oseltamivir resistance. Influenza A(H3N2) viruses belonged to subclade 3C.2a1b.2a.2 with the genetic group 2b being dominant. All 433 influenza B viruses belonged to subclade V1A.3a.2 encoding HA1 substitutions A127T, P144L, and K203R, which could be further divided into two subgroups. None of the influenza A(H3N2) and B viruses sequenced had markers of resistance to NA inhibitors. Thus, despite the continuing circulation of Omicron descendant lineages, influenza activity has resumed in full force, raising concerns about the intensity of fore coming seasonal epidemics.
Keywords: SARS CoV-2; antigenic properties; genetic analysis; hospitalization; incidence; influenza; molecular detection; monitoring; susceptibility to antivirals.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Paget J., Spreeuwenberg P., Charu V., Taylor R.J., Iuliano A.D., Bresee J., Simonsen L., Viboud C. Global mortality associated with seasonal influenza epidemics: New burden estimates and predictors from the GLaMOR Project. J. Glob. Health. 2019;9:020421. doi: 10.7189/jogh.09.020421. - DOI - PMC - PubMed
-
- Baselga-Moreno V., Trushakova S., McNeil S., Sominina A., Nunes M.C., Draganescu A., Unal S., Koul P., Kyncl J., Zhang T., et al. Influenza epidemiology and influenza vaccine effectiveness during the 2016–2017 season in the Global Influenza Hospital Surveillance Network (GIHSN) BMC Public Health. 2019;19:487. doi: 10.1186/s12889-019-6713-5. - DOI - PMC - PubMed
-
- D’Angiolella L.S., Lafranconi A., Cortesi P.A., Rota S., Cesana G., Mantovani L.G. Costs and effectiveness of influenza vaccination: A systematic review. Ann. Ist. Super. Sanita. 2018;54:49–57. - PubMed
-
- Ampofo W.K., Baylor N., Cobey S., Cox N.J., Daves S., Edwards S., Ferguson N., Grohmann G., Hay A., Katz J., et al. Improving influenza vaccine virus selection: Report of a WHO informal consultation held at WHO headquarters, Geneva, Switzerland, 14–16 June 2010. Influenza Other Respir. Viruses. 2012;6:142–152. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

