dRFEtools: dynamic recursive feature elimination for omics
- PMID: 37632789
- PMCID: PMC10471895
- DOI: 10.1093/bioinformatics/btad513
dRFEtools: dynamic recursive feature elimination for omics
Abstract
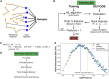
Motivation: Advances in technology have generated larger omics datasets with potential applications for machine learning. In many datasets, however, cost and limited sample availability result in an excessively higher number of features as compared to observations. Moreover, biological processes are associated with networks of core and peripheral genes, while traditional feature selection approaches capture only core genes.
Results: To overcome these limitations, we present dRFEtools that implements dynamic recursive feature elimination (RFE), reducing computational time with high accuracy compared to standard RFE, expanding dynamic RFE to regression algorithms, and outputting the subsets of features that hold predictive power with and without peripheral features. dRFEtools integrates with scikit-learn (the popular Python machine learning platform) and thus provides new opportunities for dynamic RFE in large-scale omics data while enhancing its interpretability.
Availability and implementation: dRFEtools is freely available on PyPI at https://pypi.org/project/drfetools/ or on GitHub https://github.com/LieberInstitute/dRFEtools, implemented in Python 3, and supported on Linux, Windows, and Mac OS.
© The Author(s) 2023. Published by Oxford University Press.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
References
-
- Nguyen H-N, Ohn S-Y. dRFE: dynamic recursive feature elimination for gene identification based on random forest. In: International conference on neural information processing. 2006; 4234.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

