SART3, regulated by p53, is a biomarker for diagnosis, prognosis and immune infiltration in hepatocellular carcinoma
- PMID: 37632835
- PMCID: PMC10496991
- DOI: 10.18632/aging.204978
SART3, regulated by p53, is a biomarker for diagnosis, prognosis and immune infiltration in hepatocellular carcinoma
Abstract
Objective: This study aimed to investigate the role of squamous cell carcinoma antigen recognized by T cells 3 (SART3) in hepatocellular carcinoma (HCC).
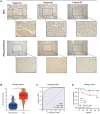
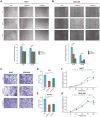
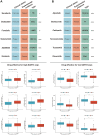
Methods: SART3 expression and prognostic value were analyzed in TCGA and GEO datasets. The diagnostic value and prognostic significance of SART3 were determined using immunohistochemistry in the Guangxi cohort. The whole-exome mutation spectrum of SART3 was analyzed in high and low expression groups in both TCGA and Guangxi cohorts. The biological functions of the SART3 gene were validated through in vitro experiments using small interfering RNA technology to downregulate SART3 expression in HCC cell lines.
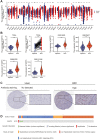
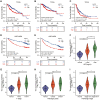
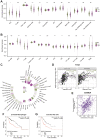
Results: SART3 expression was significantly higher in HCC tissues than in adjacent noncancerous liver tissues in TCGA, GEO and Guangxi cohorts. High expression of SART3 was significantly associated with poor prognosis in HCC patients. In TCGA and Guangxi cohorts, the expression of SART3 in the TP53 mutation group was significantly higher than that in the non-mutation group. Downregulation of SART3 expression significantly inhibited the migration and proliferation of HCC cells. SART3 may be involved mainly in immune infiltration of Th2 cells and macrophages in HCC. Additionally, SART3 can upregulate the expression of immune checkpoints (PD-L1 and TIM-3) and predict potential therapeutic agents for HCC.
Conclusion: The findings of this study demonstrate the diagnostic and prognostic value of SART3 in HCC. SART3 may be associated with immune infiltration of Th2 cells and macrophages in HCC, highlighting its potential role in the development and progression of HCC.
Keywords: TP53 mutation; hepatocellular carcinoma (HCC); immune checkpoint; immune infiltration; prognosis; squamous cell carcinoma antigen recognized by T cells 3 (SART3).
Conflict of interest statement
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

