PINK1, Keap1, and Rtnl1 regulate selective clearance of endoplasmic reticulum during development
- PMID: 37633267
- PMCID: PMC10530463
- DOI: 10.1016/j.cell.2023.08.008
PINK1, Keap1, and Rtnl1 regulate selective clearance of endoplasmic reticulum during development
Abstract
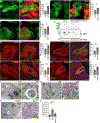
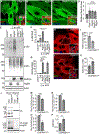
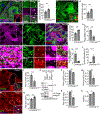
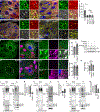
Selective clearance of organelles, including endoplasmic reticulum (ER) and mitochondria, by autophagy plays an important role in cell health. Here, we describe a developmentally programmed selective ER clearance by autophagy. We show that Parkinson's disease-associated PINK1, as well as Atl, Rtnl1, and Trp1 receptors, regulate ER clearance by autophagy. The E3 ubiquitin ligase Parkin functions downstream of PINK1 and is required for mitochondrial clearance while having the opposite function in ER clearance. By contrast, Keap1 and the E3 ubiquitin ligase Cullin3 function downstream of PINK1 to regulate ER clearance by influencing Rtnl1 and Atl. PINK1 regulates a change in Keap1 localization and Keap1-dependent ubiquitylation of the ER-phagy receptor Rtnl1 to facilitate ER clearance. Thus, PINK1 regulates the selective clearance of ER and mitochondria by influencing the balance of Keap1- and Parkin-dependent ubiquitylation of substrates that determine which organelle is removed by autophagy.
Keywords: Drosophila; ER-phagy; Keap1; PINK1; Parkin; Rtnl1.
Copyright © 2023 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

