Plasma proteomic profiling discovers molecular features associated with upper tract urothelial carcinoma
- PMID: 37633276
- PMCID: PMC10518597
- DOI: 10.1016/j.xcrm.2023.101166
Plasma proteomic profiling discovers molecular features associated with upper tract urothelial carcinoma
Abstract
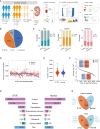
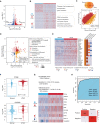
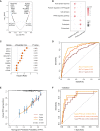
Upper tract urothelial carcinoma (UTUC) is often diagnosed late and exhibits poor prognosis. Limited data are available on potential non-invasive biomarkers for disease monitoring. Here, we investigate the proteomic profile of plasma in 362 UTUC patients and 239 healthy controls. We present an integrated tissue-plasma proteomic approach to infer the signature proteins for identifying patients with muscle-invasive UTUC. We discover a protein panel that reflects lymph node metastasis, which is of interest in identifying UTUC patients with high risk and poor prognosis. We also identify a ten-protein classifier and establish a progression clock predicting progression-free survival of UTUC patients. Finally, we further validate the signature proteins by parallel reaction monitoring assay in an independent cohort. Collectively, this study portrays the plasma proteomic landscape of a UTUC cohort and provides a valuable resource for further biological and diagnostic research in UTUC.
Keywords: DIA; mass spectrometry; plasma proteomics; prognosis; upper tract urothelial carcinoma.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures





References
-
- Rouprêt M., Babjuk M., Burger M., Capoun O., Cohen D., Compérat E.M., Cowan N.C., Dominguez-Escrig J.L., Gontero P., Hugh Mostafid A., et al. European association of Urology guidelines on upper urinary tract urothelial carcinoma: 2020 update. Eur. Urol. 2021;79:62–79. doi: 10.1016/j.eururo.2020.05.042. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

