Identified and potential internalization signals involved in trafficking and regulation of Na+/K+ ATPase activity
- PMID: 37634170
- PMCID: PMC11254989
- DOI: 10.1007/s11010-023-04831-y
Identified and potential internalization signals involved in trafficking and regulation of Na+/K+ ATPase activity
Abstract
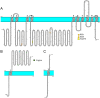
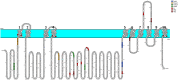
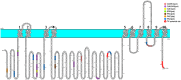
The sodium-potassium pump (NKA) or Na+/K+ ATPase consumes around 30-40% of the total energy expenditure of the animal cell on the generation of the sodium and potassium electrochemical gradients that regulate various electrolyte and nutrient transport processes. The vital role of this protein entails proper spatial and temporal regulation of its activity through modulatory mechanisms involving its expression, localization, enzymatic activity, and protein-protein interactions. The residence of the NKA at the plasma membrane is compulsory for its action as an antiporter. Despite the huge body of literature reporting on its trafficking between the cell membrane and intracellular compartments, the mechanisms controlling the trafficking process are by far the least understood. Among the molecular determinants of the plasma membrane proteins trafficking are intrinsic sequence-based endocytic motifs. In this review, we (i) summarize previous reports linking the regulation of Na+/K+ ATPase trafficking and/or plasma membrane residence to its activity, with particular emphasis on the endocytic signals in the Na+/K+ ATPase alpha-subunit, (ii) map additional potential internalization signals within Na+/K+ ATPase catalytic alpha-subunit, based on canonical and noncanonical endocytic motifs reported in the literature, (iii) pinpoint known and potential phosphorylation sites associated with NKA trafficking, (iv) highlight our recent studies on Na+/K+ ATPase trafficking and PGE2-mediated Na+/K+ ATPase modulation in intestine, liver, and kidney cells.
Keywords: Endocytic motifs; Internalization; K+ ATPase; Na+; PGE2; Phosphorylation; Trafficking.
© 2023. The Author(s).
Conflict of interest statement
The authors have no conflict of interest to declare.
Figures
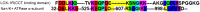
Similar articles
-
Regulation of Neuronal Na+/K+-ATPase by Specific Protein Kinases and Protein Phosphatases.J Neurosci. 2019 Jul 10;39(28):5440-5451. doi: 10.1523/JNEUROSCI.0265-19.2019. Epub 2019 May 13. J Neurosci. 2019. PMID: 31085608 Free PMC article.
-
G protein-coupled receptors regulate Na+,K+-ATPase activity and endocytosis by modulating the recruitment of adaptor protein 2 and clathrin.Proc Natl Acad Sci U S A. 2000 Mar 28;97(7):3242-7. doi: 10.1073/pnas.97.7.3242. Proc Natl Acad Sci U S A. 2000. PMID: 10716725 Free PMC article.
-
Localization of intracellular compartments that exchange Na,K-ATPase molecules with the plasma membrane in a hormone-dependent manner.Br J Pharmacol. 2007 Aug;151(7):1006-13. doi: 10.1038/sj.bjp.0707304. Epub 2007 May 29. Br J Pharmacol. 2007. PMID: 17533417 Free PMC article.
-
Na,K-ATPase regulation in skeletal muscle.Am J Physiol Endocrinol Metab. 2016 Jul 1;311(1):E1-E31. doi: 10.1152/ajpendo.00539.2015. Epub 2016 May 10. Am J Physiol Endocrinol Metab. 2016. PMID: 27166285 Review.
-
Na(+),K (+)-ATPase as a docking station: protein-protein complexes of the Na(+),K (+)-ATPase.Cell Mol Life Sci. 2013 Jan;70(2):205-22. doi: 10.1007/s00018-012-1039-9. Epub 2012 Jun 14. Cell Mol Life Sci. 2013. PMID: 22695678 Free PMC article. Review.
Cited by
-
Na+/K+-ATPase: a multifunctional target in type 2 diabetes and pancreatic islets.Front Immunol. 2025 Feb 19;16:1555310. doi: 10.3389/fimmu.2025.1555310. eCollection 2025. Front Immunol. 2025. PMID: 40046060 Free PMC article. Review.
-
Integrative transcriptomics and metabolomics reveal neuroendocrine-lipid crosstalk and adenosine signaling in broiler under heat stress.BMC Genomics. 2025 Jul 29;26(1):699. doi: 10.1186/s12864-025-11881-7. BMC Genomics. 2025. PMID: 40731384 Free PMC article.
References
-
- Mobasheri A, Avila J, Cózar-Castellano I, Brownleader MD, Trevan M, Francis MJ, Lamb JF, Martín-Vasallo P. Na+, K+-ATPase isozyme diversity; comparative biochemistry and physiological implications of novel functional interactions. Biosci Rep. 2000;20(2):51–91. doi: 10.1023/a:1005580332144. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

