Hypomyelination, hypodontia and craniofacial abnormalities in a Polr3b mouse model of leukodystrophy
- PMID: 37635302
- PMCID: PMC10690025
- DOI: 10.1093/brain/awad249
Hypomyelination, hypodontia and craniofacial abnormalities in a Polr3b mouse model of leukodystrophy
Abstract
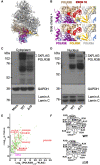
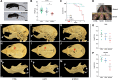
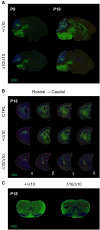
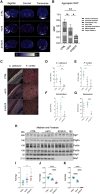
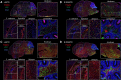
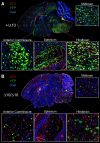
RNA polymerase III (Pol III)-related hypomyelinating leukodystrophy (POLR3-HLD), also known as 4H leukodystrophy, is a severe neurodegenerative disease characterized by the cardinal features of hypomyelination, hypodontia and hypogonadotropic hypogonadism. POLR3-HLD is caused by biallelic pathogenic variants in genes encoding Pol III subunits. While approximately half of all patients carry mutations in POLR3B encoding the RNA polymerase III subunit B, there is no in vivo model of leukodystrophy based on mutation of this Pol III subunit. Here, we determined the impact of POLR3BΔ10 (Δ10) on Pol III in human cells and developed and characterized an inducible/conditional mouse model of leukodystrophy using the orthologous Δ10 mutation in mice. The molecular mechanism of Pol III dysfunction was determined in human cells by affinity purification-mass spectrometry and western blot. Postnatal induction with tamoxifen induced expression of the orthologous Δ10 hypomorph in triple transgenic Pdgfrα-Cre/ERT; R26-Stopfl-EYFP; Polr3bfl mice. CNS and non-CNS features were characterized using a variety of techniques including microCT, ex vivo MRI, immunofluorescence, immunohistochemistry, spectral confocal reflectance microscopy and western blot. Lineage tracing and time series analysis of oligodendrocyte subpopulation dynamics based on co-labelling with lineage-specific and/or proliferation markers were performed. Proteomics suggested that Δ10 causes a Pol III assembly defect, while western blots demonstrated reduced POLR3BΔ10 expression in the cytoplasm and nucleus in human cells. In mice, postnatal Pdgfrα-dependent expression of the orthologous murine mutant protein resulted in recessive phenotypes including severe hypomyelination leading to ataxia, tremor, seizures and limited survival, as well as hypodontia and craniofacial abnormalities. Hypomyelination was confirmed and characterized using classic methods to quantify myelin components such as myelin basic protein and lipids, results which agreed with those produced using modern methods to quantify myelin based on the physical properties of myelin membranes. Lineage tracing uncovered the underlying mechanism for the hypomyelinating phenotype: defective oligodendrocyte precursor proliferation and differentiation resulted in a failure to produce an adequate number of mature oligodendrocytes during postnatal myelinogenesis. In summary, we characterized the Polr3bΔ10 mutation and developed an animal model that recapitulates features of POLR3-HLD caused by POLR3B mutations, shedding light on disease pathogenesis, and opening the door to the development of therapeutic interventions.
Keywords: 4H leukodystrophy; POLR3-related leukodystrophy; POLR3B; mouse model; myelinogenesis; neurodevelopment.
© The Author(s) 2023. Published by Oxford University Press on behalf of the Guarantors of Brain.
Conflict of interest statement
G.B. was a consultant for Passage Bio Inc (2020-2022) and Ionis (2019). She is/was a site investigator for the Alexander’s disease trial of Ionis (2021–present), Metachromatic leukodystrophy of Shire/Takeda (2020–2021), Krabbe and GM1 gene therapy trials of Passage Bio (2021–present), GM1 natural history study from the University of Pennsylvania sponsored by Passage Bio (2021–present) and Adrenoleukodystrophy/Hematopoietic stem cell transplantation natural history study of Bluebird Bio (2019), a site sub-investigator for the MPS II gene therapy trial of Regenxbio (2021–present) and the MPS II clinical trial of Denali (2022–present). She has received unrestricted educational grants from Takeda (2021–2022). The other authors have no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

