Juno and CD9 protein network organization in oolemma of mouse oocyte
- PMID: 37635875
- PMCID: PMC10450504
- DOI: 10.3389/fcell.2023.1110681
Juno and CD9 protein network organization in oolemma of mouse oocyte
Abstract
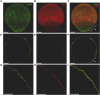
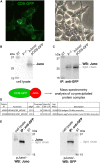
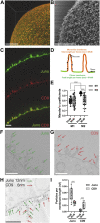
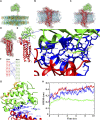
Juno and CD9 protein, expressed in oolemma, are known to be essential for sperm-oocyte binding and fusion. Although evidence exists that these two proteins cooperate, their interaction has not yet been demonstrated. Here in, we present Juno and CD9 mutual localization over the surface of mouse metaphase II oocytes captured using the 3D STED super-resolution technique. The precise localization of examined proteins was identified in different compartments of oolemma such as the microvillar membrane, planar membrane between individual microvilli, and the membrane of microvilli-free region. Observed variance in localization of Juno and CD9 was confirmed by analysis of transmission and scanning electron microscopy images, which showed a significant difference in the presence of proteins between selected membrane compartments. Colocalization analysis of super-resolution images based on Pearson's correlation coefficient supported evidence of Juno and CD9 mutual position in the oolemma, which was identified by proximity ligation assay. Importantly, the interaction between Juno and CD9 was detected by co-immunoprecipitation and mass spectrometry in HEK293T/17 transfected cell line. For better understanding of experimental data, mouse Juno and CD9 3D structure were prepared by comparative homology modelling and several protein-protein flexible sidechain dockings were performed using the ClusPro server. The dynamic state of the proteins was studied in real-time at atomic level by molecular dynamics (MD) simulation. Docking and MD simulation predicted Juno-CD9 interactions and stability also suggesting an interactive mechanism. Using the multiscale approach, we detected close proximity of Juno and CD9 within microvillar oolemma however, not in the planar membrane or microvilli-free region. Our findings show yet unidentified Juno and CD9 interaction within the mouse oolemma protein network prior to sperm attachment. These results suggest that a Juno and CD9 interactive network could assist in primary Juno binding to sperm Izumo1 as a prerequisite to subsequent gamete membrane fusion.
Keywords: CD9; Juno; MD simulation; STED; docking; oocyte; oolemma compartments; protein interaction.
Copyright © 2023 Frolikova, Sur, Novotny, Blazikova, Vondrakova, Simonik, Ded, Valaskova, Koptasikova, Benda, Postlerova, Horvath and Komrskova.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
LinkOut - more resources
Full Text Sources

