Spf1 and Ste24: quality controllers of transmembrane protein topology in the eukaryotic cell
- PMID: 37635876
- PMCID: PMC10456885
- DOI: 10.3389/fcell.2023.1220441
Spf1 and Ste24: quality controllers of transmembrane protein topology in the eukaryotic cell
Abstract
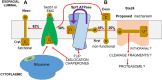
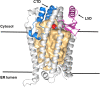
DNA replication, transcription, and translation in eukaryotic cells occur with decreasing but still high fidelity. In contrast, for the estimated 33% of the human proteome that is inserted as transmembrane (TM) proteins, insertion with a non-functional inverted topology is frequent. Correct topology is essential for function and trafficking to appropriate cellular compartments and is controlled principally by responses to charged residues within 15 residues of the inserted TM domain (TMD); the flank with the higher positive charge remains in the cytosol (inside), following the positive inside rule (PIR). Yeast (Saccharomyces cerevisiae) mutants that increase insertion contrary to the PIR were selected. Mutants with strong phenotypes were found only in SPF1 and STE24 (human cell orthologs are ATP13A1 and ZMPSte24) with, at the time, no known relevant functions. Spf1/Atp13A1 is now known to dislocate to the cytosol TM proteins inserted contrary to the PIR, allowing energy-conserving reinsertion. We hypothesize that Spf1 and Ste24 both recognize the short, positively charged ER luminal peptides of TM proteins inserted contrary to the PIR, accepting these peptides into their large membrane-spanning, water-filled cavities through interaction with their many interior surface negative charges. While entry was demonstrated for Spf1, no published evidence directly demonstrates substrate entry to the Ste24 cavity, internal access to its zinc metalloprotease (ZMP) site, or active withdrawal of fragments, which may be essential for function. Spf1 and Ste24 comprise a PIR quality control system that is conserved in all eukaryotes and presumably evolved in prokaryotic progenitors as they gained differentiated membrane functions. About 75% of the PIR is imposed by this quality control system, which joins the UPR, ERAD, and autophagy (ER-phagy) in coordinated, overlapping quality control of ER protein function.
Keywords: positive inside rule; quality control; topology; topology error recognition; transmembrane proteins.
Copyright © 2023 Tipper and Harley.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
Publication types
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

