Proof of concept of using a membrane-sensing peptide for sEVs affinity-based isolation
- PMID: 37636002
- PMCID: PMC10457001
- DOI: 10.3389/fbioe.2023.1238898
Proof of concept of using a membrane-sensing peptide for sEVs affinity-based isolation
Abstract
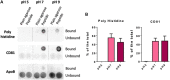
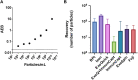
Introduction: One main limitation in biomarker studies using EVs is the lack of a suitable isolation method rendering high yield and purity samples in a quick and easily standardized procedure. Here we report an affinity isolation method with a membrane-sensing peptide (MSP) derived from bradykinin. Methods: We designed a protocol based on agarose beads carrying cation chelates to specifically bind to the 6His-tagged membrane-sensing peptide. This approach presents several advantages: 1) cation-carrying agaroses are widely used and standardized for His-tagged protein isolation, 2) the affinity protocol can be performed in small volumes, feasible and manageable for clinical routine and 3) elution with imidazole or EDTA allows a gentle and easy recovery without EV damage, facilitating subsequent characterization and functional analyses. Results: The optimized final procedure incubates 0.5 mg of peptide for 10 min with 10 µL of Long-arm Cobalt agarose before an overnight incubation with concentrated cell conditioned medium. EV downstream analyses can be directly performed on the agarose beads adding lysis or nucleic-acid extraction buffers, or gently eluted with imidazole or EDTA, rendering a fully competent EV preparation. Discussion: This new isolation methodology is based on the recognition of general membrane characteristics independent of surface markers. It is thus unbiased and can be used in any species EV sample, even in samples from animal or plant species against which no suitable antibodies exist. Being an affinity method, the sample handling protocol is very simple, less time-consuming, does not require specialized equipment and can be easily introduced in a clinical automated routine. We demonstrated the high purity and yield of the method in comparison with other commercially available kits. This method can also be scale up or down, with the possibility of analyzing very low amounts of sample, and it is compatible with any downstream analyses thanks to the gentle elution procedure.
Keywords: affinity; agarose beads; extracellular vesicles; isolation; peptide.
Copyright © 2023 Benayas, Morales, Gori, Strada, Gagni, Frigerio, Egea, Armisén, Cretich and Yáñez-Mó.
Conflict of interest statement
CE is general manager of ABT, PA is Business and technical development manager at ABT and BB is employed by ABT in the context of an industrial doctorate. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures







References
-
- Campos-Silva C., Cáceres-Martell Y., López-Cobo S., Rodriguez M. J., Jara R., Yáñez-Mó M., et al. (2021). “An immunocapture-based assay for detecting multiple antigens in melanoma-derived extracellular vesicles,” in Melanoma: Methods and protocols. Editor Hargadon E. K. M. (USA: Springer; ), 323–344. 10.1007/978-1-0716-1205-7_24 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

