Intracellular peptides in SARS-CoV-2-infected patients
- PMID: 37636076
- PMCID: PMC10448160
- DOI: 10.1016/j.isci.2023.107542
Intracellular peptides in SARS-CoV-2-infected patients
Abstract
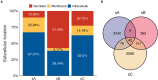
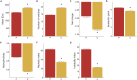
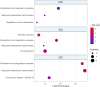
Intracellular peptides (InPeps) generated by the orchestrated action of the proteasome and intracellular peptidases have biological and pharmacological significance. Here, human plasma relative concentration of specific InPeps was compared between 175 patients infected with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), and 45 SARS-CoV-2 non-infected patients; 2,466 unique peptides were identified, of which 67% were InPeps. The results revealed differences of a specific group of peptides in human plasma comparing non-infected individuals to patients infected by SARS-CoV-2, following the results of the semi-quantitative analyses by isotope-labeled electrospray mass spectrometry. The protein-protein interactions networks enriched pathways, drawn by genes encoding the proteins from which the peptides originated, revealed the presence of the coronavirus disease/COVID-19 network solely in the group of patients fatally infected by SARS-CoV-2. Thus, modulation of the relative plasma levels of specific InPeps could be employed as a predictive tool for disease outcome.
Keywords: Microbiology; Public health; Virology.
© 2023 The Author(s).
Conflict of interest statement
All authors declare no conflict of interest.
Figures


References
-
- Javitt A., Shmueli M.D., Kramer M.P., Kolodziejczyk A.A., Cohen I.J., Radomir L., Sheban D., Kamer I., Litchfield K., Bab-Dinitz E., et al. The proteasome regulator PSME4 modulates proteasome activity and antigen diversity to abrogate antitumor immunity in NSCLC. Nat. Cancer. 2023;4:629–647. doi: 10.1038/s43018-023-00557-4. - DOI - PubMed
-
- Captur G., Moon J.C., Topriceanu C.-C., Joy G., Swadling L., Hallqvist J., Doykov I., Patel N., Spiewak J., Baldwin T., et al. Plasma proteomic signature predicts who will get persistent symptoms following SARS-CoV-2 infection. EBioMedicine. 2022;85:104293. doi: 10.1016/j.ebiom.2022.104293. - DOI - PMC - PubMed
-
- Staessen J.A., Wendt R., Yu Y.L., Kalbitz S., Thijs L., Siwy J., Raad J., Metzger J., Neuhaus B., Papkalla A., et al. Predictive performance and clinical application of COV50, a urinary proteomic biomarker in early COVID-19 infection: a prospective multicentre cohort study. Lancet. Digit. Health. 2022;4:e727–e737. doi: 10.1016/s2589-7500(22)00150-9. - DOI - PMC - PubMed
-
- Palanski B.A., Weng N., Zhang L., Hilmer A.J., Fall L.A., Swaminathan K., Jabri B., Sousa C., Fernandez-Becker N.Q., Khosla C., Elias J.E. An efficient urine peptidomics workflow identifies chemically defined dietary gluten peptides from patients with celiac disease. Nat. Commun. 2022;13:888. doi: 10.1038/s41467-022-28353-1. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

