Interferome signature dynamics during the anti-dengue immune response: a systems biology characterization
- PMID: 37638052
- PMCID: PMC10449254
- DOI: 10.3389/fimmu.2023.1243516
Interferome signature dynamics during the anti-dengue immune response: a systems biology characterization
Abstract
Dengue virus (DENV) infection manifests as a febrile illness with three distinct phases: early acute, late acute, and convalescent. Dengue can result in clinical manifestations with different degrees of severity, dengue fever, dengue hemorrhagic fever, and dengue shock syndrome. Interferons (IFNs) are antiviral cytokines central to the anti-DENV immune response. Notably, the distinct global signature of type I, II, and III interferon-regulated genes (the interferome) remains uncharacterized in dengue patients to date. Therefore, we performed an in-depth cross-study for the integrative analysis of transcriptome data related to DENV infection. Our systems biology analysis shows that the anti-dengue immune response is characterized by the modulation of numerous interferon-regulated genes (IRGs) enriching, for instance, cytokine-mediated signaling (e.g., type I and II IFNs) and chemotaxis, which is then followed by a transcriptional wave of genes associated with cell cycle, also regulated by the IFN cascade. The adjunct analysis of disease stratification potential, followed by a transcriptional meta-analysis of the interferome, indicated genes such as IFI27, ISG15, and CYBRD1 as potential suitable biomarkers of disease severity. Thus, this study characterizes the landscape of the interferome signature in DENV infection, indicating that interferome dynamics are a crucial and central part of the anti-dengue immune response.
Keywords: DENV; dengue; interferome; interferon; transcriptome.
Copyright © 2023 Usuda, Plaça, Fonseca, Marques, Filgueiras, Chaves, Adri, Torrentes-Carvalho, Hirata, Freire, Catar, Cabral-Miranda, Schimke, Moll and Cabral-Marques.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


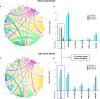
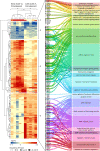


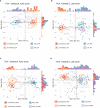

References
-
- World Health Organization . Ending the neglect to attain the Sustainable Development Goals: A road map for neglected tropical diseases 2021–2030. Overview (2021). Available at: https://www.who.int/publications-detail-redirect/WHO-UCN-NTD-2020.01 (Accessed July 1, 2021).
-
- World Health Organization, Special Programme for Research and Training in Tropical Diseases . Dengue: guidelines for diagnosis, treatment, prevention, and control. New. Geneva: World Health Organization; (2009). 147 p.
-
- Sun P, García J, Comach G, Vahey MT, Wang Z, Forshey BM, et al. Sequential waves of gene expression in patients with clinically defined dengue illnesses reveal subtle disease phases and predict disease severity. PloS Negl Trop Dis (2013) 7:e2298. doi: 10.1371/journal.pntd.0002298 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

