Role of the CagY antenna projection in Helicobacter pylori Cag type IV secretion system activity
- PMID: 37638724
- PMCID: PMC10501215
- DOI: 10.1128/iai.00150-23
Role of the CagY antenna projection in Helicobacter pylori Cag type IV secretion system activity
Abstract
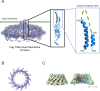
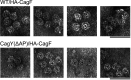
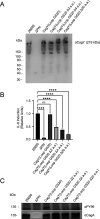
Helicobacter pylori strains containing the cag pathogenicity island (PAI) are associated with the development of gastric adenocarcinoma and peptic ulcer disease. The cag PAI encodes a secreted effector protein (CagA) and a type IV secretion system (Cag T4SS). Cag T4SS activity is required for the delivery of CagA and non-protein substrates into host cells. The Cag T4SS outer membrane core complex (OMCC) contains a channel-like domain formed by helix-loop-helix elements (antenna projections, AP) from 14 copies of the CagY protein (a VirB10 ortholog). Similar VirB10 antenna regions are present in T4SS OMCCs from multiple bacterial species and are predicted to span the outer membrane. In this study, we investigated the role of the CagY antenna region in Cag T4SS OMCC assembly and Cag T4SS function. An H. pylori mutant strain with deletion of the entire CagY AP (∆AP) retained the capacity to produce CagY and assemble an OMCC, but it lacked T4SS activity (CagA translocation and IL-8 induction in AGS gastric epithelial cells). In contrast, a mutant strain with Gly-Ser substitutions in the unstructured CagY AP loop retained Cag T4SS activity. Mutants containing CagY AP loops with shortened lengths were defective in CagA translocation and exhibited reduced IL-8-inducing activity compared to control strains. These data indicate that the CagY AP region is required for Cag T4SS activity and that Cag T4SS activity can be modulated by altering the length of the CagY AP unstructured loop.
Keywords: bacterial outer membrane protein; bacterial secretion system; gastric cancer; membrane channel; pathogenicity island.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
CagY-Dependent Regulation of Type IV Secretion in Helicobacter pylori Is Associated with Alterations in Integrin Binding.mBio. 2018 May 15;9(3):e00717-18. doi: 10.1128/mBio.00717-18. mBio. 2018. PMID: 29764950 Free PMC article.
-
CagY Is an Immune-Sensitive Regulator of the Helicobacter pylori Type IV Secretion System.Gastroenterology. 2016 Dec;151(6):1164-1175.e3. doi: 10.1053/j.gastro.2016.08.014. Epub 2016 Aug 26. Gastroenterology. 2016. PMID: 27569724 Free PMC article.
-
Helicobacter pylori CagA and Cag type IV secretion system activity have key roles in triggering gastric transcriptional and proteomic alterations.Infect Immun. 2025 Apr 8;93(4):e0059524. doi: 10.1128/iai.00595-24. Epub 2025 Mar 6. Infect Immun. 2025. PMID: 40047510 Free PMC article.
-
The Helicobacter pylori Type IV Secretion System Encoded by the cag Pathogenicity Island: Architecture, Function, and Signaling.Curr Top Microbiol Immunol. 2017;413:187-220. doi: 10.1007/978-3-319-75241-9_8. Curr Top Microbiol Immunol. 2017. PMID: 29536360 Review.
-
Composition, structure and function of the Helicobacter pylori cag pathogenicity island encoded type IV secretion system.Future Microbiol. 2015;10(6):955-65. doi: 10.2217/fmb.15.32. Future Microbiol. 2015. PMID: 26059619 Free PMC article. Review.
Cited by
-
A Proposal for a Consolidated Structural Model of the CagY Protein of Helicobacter pylori.Int J Mol Sci. 2023 Nov 26;24(23):16781. doi: 10.3390/ijms242316781. Int J Mol Sci. 2023. PMID: 38069104 Free PMC article.
-
Subdomains of the Helicobacter pylori Cag T4SS outer membrane core complex exhibit structural independence.Life Sci Alliance. 2024 Apr 17;7(6):e202302560. doi: 10.26508/lsa.202302560. Print 2024 Jun. Life Sci Alliance. 2024. PMID: 38631913 Free PMC article.
-
The Helicobacter pylori cag pathogenicity island as a determinant of gastric cancer risk.Gut Microbes. 2024 Jan-Dec;16(1):2314201. doi: 10.1080/19490976.2024.2314201. Epub 2024 Feb 23. Gut Microbes. 2024. PMID: 38391242 Free PMC article. Review.
References
-
- Hooi JKY, Lai WY, Ng WK, Suen MMY, Underwood FE, Tanyingoh D, Malfertheiner P, Graham DY, Wong VWS, Wu JCY, Chan FKL, Sung JJY, Kaplan GG, Ng SC. 2017. Global prevalence of Helicobacter pylori infection: systematic review and meta-analysis. Gastroenterology 153:420–429. doi:10.1053/j.gastro.2017.04.022 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

