Sec22b and Stx4 Depletion Has No Major Effect on Cross-Presentation of PLGA Microsphere-Encapsulated Antigen and a Synthetic Long Peptide In Vitro
- PMID: 37638825
- PMCID: PMC10592162
- DOI: 10.4049/jimmunol.2200473
Sec22b and Stx4 Depletion Has No Major Effect on Cross-Presentation of PLGA Microsphere-Encapsulated Antigen and a Synthetic Long Peptide In Vitro
Abstract
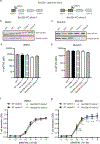
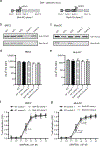
The induction of CTL responses by vaccines is important to combat infectious diseases and cancer. Biodegradable poly(lactic-co-glycolic acid) (PLGA) microspheres and synthetic long peptides are efficiently internalized by professional APCs and prime CTL responses after cross-presentation of Ags on MHC class I molecules. Specifically, they mainly use the cytosolic pathway of cross-presentation that requires endosomal escape, proteasomal processing, and subsequent MHC class I loading of Ags in the endoplasmic reticulum (ER) and/or the endosome. The vesicle SNARE protein Sec22b has been described as important for this pathway by mediating vesical trafficking for the delivery of ER-derived proteins to the endosome. As this function has also been challenged, we investigated the role of Sec22b in cross-presentation of the PLGA microsphere-encapsulated model Ag OVA and a related synthetic long peptide. Using CRISPR/Cas9-mediated genome editing, we generated Sec22b knockouts in two murine C57BL/6-derived APC lines and found no evidence for an essential role of Sec22b. Although pending experimental evidence, the target SNARE protein syntaxin 4 (Stx4) has been suggested to promote cross-presentation by interacting with Sec22b for the fusion of ER-derived vesicles with the endosome. In the current study, we show that, similar to Sec22b, Stx4 knockout in murine APCs had very limited effects on cross-presentation under the conditions tested. This study contributes to characterizing cross-presentation of two promising Ag delivery systems and adds to the discussion about the role of Sec22b/Stx4 in related pathways. Our data point toward SNARE protein redundancy in the cytosolic pathway of cross-presentation.
Copyright © 2023 by The American Association of Immunologists, Inc.
Figures



References
-
- Joffre OP, Segura E, Savina A, and Amigorena S. 2012. Cross-presentation by dendritic cells. Nat Rev Immunol 12: 557–569. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

