GPRC5C drives branched-chain amino acid metabolism in leukemogenesis
- PMID: 37639313
- PMCID: PMC10761356
- DOI: 10.1182/bloodadvances.2023010460
GPRC5C drives branched-chain amino acid metabolism in leukemogenesis
Abstract
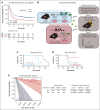
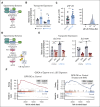
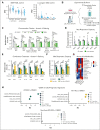
Leukemia stem cells (LSCs) share numerous features with healthy hematopoietic stem cells (HSCs). G-protein coupled receptor family C group 5 member C (GPRC5C) is a regulator of HSC dormancy. However, GPRC5C functionality in acute myeloid leukemia (AML) is yet to be determined. Within patient AML cohorts, high GPRC5C levels correlated with poorer survival. Ectopic Gprc5c expression increased AML aggression through the activation of NF-κB, which resulted in an altered metabolic state with increased levels of intracellular branched-chain amino acids (BCAAs). This onco-metabolic profile was reversed upon loss of Gprc5c, which also abrogated the leukemia-initiating potential. Targeting the BCAA transporter SLC7A5 with JPH203 inhibited oxidative phosphorylation and elicited strong antileukemia effects, specifically in mouse and patient AML samples while sparing healthy bone marrow cells. This antileukemia effect was strengthened in the presence of venetoclax and azacitidine. Our results indicate that the GPRC5C-NF-κB-SLC7A5-BCAAs axis is a therapeutic target that can compromise leukemia stem cell function in AML.
© 2023 by The American Society of Hematology. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Conflict of interest statement
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

