Genetic enhancers of partial PLK1 inhibition reveal hypersensitivity to kinetochore perturbations
- PMID: 37639469
- PMCID: PMC10491399
- DOI: 10.1371/journal.pgen.1010903
Genetic enhancers of partial PLK1 inhibition reveal hypersensitivity to kinetochore perturbations
Abstract
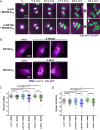
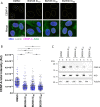
Polo-like kinase 1 (PLK1) is a serine/threonine kinase required for mitosis and cytokinesis. As cancer cells are often hypersensitive to partial PLK1 inactivation, chemical inhibitors of PLK1 have been developed and tested in clinical trials. However, these small molecule inhibitors alone are not completely effective. PLK1 promotes numerous molecular and cellular events in the cell division cycle and it is unclear which of these events most crucially depend on PLK1 activity. We used a CRISPR-based genome-wide screening strategy to identify genes whose inactivation enhances cell proliferation defects upon partial chemical inhibition of PLK1. Genes identified encode proteins that are functionally linked to PLK1 in multiple ways, most notably factors that promote centromere and kinetochore function. Loss of the kinesin KIF18A or the outer kinetochore protein SKA1 in PLK1-compromised cells resulted in mitotic defects, activation of the spindle assembly checkpoint and nuclear reassembly defects. We also show that PLK1-dependent CENP-A loading at centromeres is extremely sensitive to partial PLK1 inhibition. Our results suggest that partial inhibition of PLK1 compromises the integrity and function of the centromere/kinetochore complex, rendering cells hypersensitive to different kinetochore perturbations. We propose that KIF18A is a promising target for combinatorial therapies with PLK1 inhibitors.
Copyright: © 2023 Normandin et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
-
- Morgan DO. The Cell Cycle: Principles of Control. London: New Science Press; 2007. 297 p.
-
- Sunkel CE, Glover DM. polo, a mitotic mutant of Drosophila displaying abnormal spindle poles. J Cell Sci. 1988;89 (Pt 1):25–38. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

