H3K4me3 remodeling induced acquired resistance through O-GlcNAc transferase
- PMID: 37639774
- PMCID: PMC10719180
- DOI: 10.1016/j.drup.2023.100993
H3K4me3 remodeling induced acquired resistance through O-GlcNAc transferase
Abstract
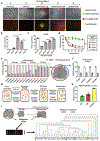
Aims: Drivers of the drug tolerant proliferative persister (DTPP) state have not been well investigated. Histone H3 lysine-4 trimethylation (H3K4me3), an active histone mark, might enable slow cycling drug tolerant persisters (DTP) to regain proliferative capacity. This study aimed to determine H3K4me3 transcriptionally active sites identifying a key regulator of DTPPs.
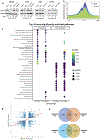
Methods: Deploying a model of adaptive cancer drug tolerance, H3K4me3 ChIP-Seq data of DTPPs guided identification of top transcription factor binding motifs. These suggested involvement of O-linked N-acetylglucosamine transferase (OGT), which was confirmed by metabolomics analysis and biochemical assays. OGT impact on DTPPs and adaptive resistance was explored in vitro and in vivo.
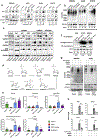
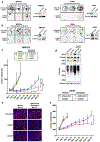
Results: H3K4me3 remodeling was widespread in CPG island regions and DNA binding motifs associated with O-GlcNAc marked chromatin. Accordingly, we observed an upregulation of OGT, O-GlcNAc and its binding partner TET1 in chronically treated cancer cells. Inhibition of OGT led to loss of H3K4me3 and downregulation of genes contributing to drug resistance. Genetic ablation of OGT prevented acquired drug resistance in in vivo models. Upstream of OGT, we identified AMPK as an actionable target. AMPK activation by acetyl salicylic acid downregulated OGT with similar effects on delaying acquired resistance.
Conclusion: Our findings uncover a fundamental mechanism of adaptive drug resistance that governs cancer cell reprogramming towards acquired drug resistance, a process that can be exploited to improve response duration and patient outcomes.
Keywords: Acquired drug resistance; Adaptive cancer drug resistance; Cancer persisters; Cellular reprogramming; Epigenetics; H3K4me3; Metabolism; OGT; TET1.
Copyright © 2023 The Authors. Published by Elsevier Ltd.. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no competing financial interests or personal relationships that influence the work reported in this paper.
Figures

References
-
- Ahronian LG, Sennott EM, Van Allen EM, Wagle N, Kwak EL, Faris JE, Godfrey JT, Nishimura K, Lynch KD, Mermel CH, Lockerman EL, Kalsy A, Gurski JM Jr., Bahl S, Anderka K, Green LM, Lennon NJ, Huynh TG, Mino-Kenudson M, Getz G, Dias-Santagata D, Iafrate AJ, Engelman JA, Garraway LA, Corcoran RB, 2015. Clinical acquired resistance to RAF inhibitor combinations in BRAF-mutant colorectal cancer through MAPK pathway alterations. Cancer Discov. 5 (4), 358–367. - PMC - PubMed
-
- Buescher JM, Moco S, Sauer U, Zamboni N, 2010. Ultrahigh performance liquid chromatography– tandem mass spectrometry method for fast and robust quantification of anionic and aromatic metabolites. Analytical chemistry 82 (11), 4403–4412. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

