Engaging an HIV vaccine target through the acquisition of low B cell affinity
- PMID: 37640732
- PMCID: PMC10462694
- DOI: 10.1038/s41467-023-40918-2
Engaging an HIV vaccine target through the acquisition of low B cell affinity
Abstract
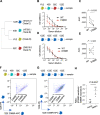
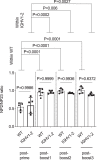
Low affinity is common for germline B cell receptors (BCR) seeding development of broadly neutralizing antibodies (bnAbs) that engage hypervariable viruses, including HIV. Antibody affinity selection is also non-homogenizing, insuring the survival of low affinity B cell clones. To explore whether this provides a natural window for expanding human B cell lineages against conserved vaccine targets, we deploy transgenic mice mimicking human antibody diversity and somatic hypermutation (SHM) and immunize with simple monomeric HIV glycoprotein envelope immunogens. We report an immunization regimen that focuses B cell memory upon the conserved CD4 binding site (CD4bs) through both conventional affinity maturation and reproducible expansion of low affinity BCR clones with public patterns in SHM. In the latter instance, SHM facilitates target acquisition by decreasing binding strength. This suggests that permissive B cell selection enables the discovery of antibody epitopes, in this case an HIV bnAb site.
© 2023. Springer Nature Limited.
Conflict of interest statement
D.L. reports SAB membership for Flagship Labs 72, Tendel Therapies, and Lattice Therapeutics Inc. The remaining authors declare no other competing interests.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

