Comparison of bivalent and monovalent SARS-CoV-2 variant vaccines: the phase 2 randomized open-label COVAIL trial
- PMID: 37640860
- PMCID: PMC10504073
- DOI: 10.1038/s41591-023-02503-4
Comparison of bivalent and monovalent SARS-CoV-2 variant vaccines: the phase 2 randomized open-label COVAIL trial
Abstract
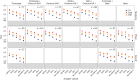
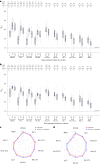
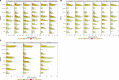
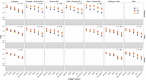
Vaccine protection against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection wanes over time, requiring updated boosters. In a phase 2, open-label, randomized clinical trial with sequentially enrolled stages at 22 US sites, we assessed safety and immunogenicity of a second boost with monovalent or bivalent variant vaccines from mRNA and protein-based platforms targeting wild-type, Beta, Delta and Omicron BA.1 spike antigens. The primary outcome was pseudovirus neutralization titers at 50% inhibitory dilution (ID50 titers) with 95% confidence intervals against different SARS-CoV-2 strains. The secondary outcome assessed safety by solicited local and systemic adverse events (AEs), unsolicited AEs, serious AEs and AEs of special interest. Boosting with prototype/wild-type vaccines produced numerically lower ID50 titers than any variant-containing vaccine against all variants. Conversely, boosting with a variant vaccine excluding prototype was not associated with decreased neutralization against D614G. Omicron BA.1 or Beta monovalent vaccines were nearly equivalent to Omicron BA.1 + prototype or Beta + prototype bivalent vaccines for neutralization of Beta, Omicron BA.1 and Omicron BA.4/5, although they were lower for contemporaneous Omicron subvariants. Safety was similar across arms and stages and comparable to previous reports. Our study shows that updated vaccines targeting Beta or Omicron BA.1 provide broadly crossprotective neutralizing antibody responses against diverse SARS-CoV-2 variants without sacrificing immunity to the ancestral strain. ClinicalTrials.gov registration: NCT05289037 .
© 2023. The Author(s).
Conflict of interest statement
L.G. has received funding from Leidos Biomedical Research. E.J.A. has received funding from Pfizer, Moderna, Janssen, GSK, Sanofi Pasteur, Micron and Regeneron (payment made to institution); and serves a safety monitoring role for Sanofi Pasteur, ACI Clinical/WCG and Kentucky Bioscience (payment made to E.J.A.). J.A. has received funding from the Division of Microbiology and Infectious Diseases, contract no. 75N93021C00012. A.R.B. has received grant funding from NIH, NIAID, Pfizer, Cyanvac and Merck (payment to institution); and has received consulting fees from Janssen and GSK (payment made to A.R.B.). L.R.B. has received grant funding from NIH, NIH/Harvard Medical School, Wellcome Trust and the Gates Foundation; and serves in a data safety monitoring role for NIH, US Food and Drug Administration (payment made to institution). She is involved in HIV and SARS-CoV-2 vaccine clinical trials conducted in collaboration with the NIH, HIV Vaccine Trials Network, COVID Vaccine Prevention Network, International AIDS Vaccine Initiative, Crucell/Janssen, Moderna, Military HIV Research Program, the Gates Foundation and Harvard Medical School. D.J.D. has received funding from Leidos Biomedical (research contract to his institution) to conduct a clinical trial. A.E. has received DMID funding via NIH subcontract to Duke University for the CIVICS Option 21 and Moderna. H.M.E.S. has received funding support from the NIH (grant no. 22CTA-DM0002). A.R.F. has received grant funding from Janssen, Pfizer, Merck, BioFire Diagnostics and CyanVac (payment made to institution); and has received consultant fees from Arrowhead, Icosavax, Moderna and GlaxoSmithKline (payable to A.R.F.). A.R.F. also reports travel meeting support from GlaxoSmithKline, and serving in a data safety monitoring role for Novavax. S.E.F. has received funding from Leidos to Saint Louis University to conduct the protocol, DMID22-0004. D.N.F. has received a contract from the CDC and is the site PI for clinical trials from Gilead, Regeneron and MetroBiothech. She is the PI on one investigator-initiated award from Gilead and served on an advisory board for Gilead in 2021. P.A.G. has received COVAIL clinical trial funding and consulting fees from Janssen Vaccines (payment made to P.A.G.). D.S.G. has received funding from NIAID/IDCRC, Henry M. Jackson Foundation Contract, and consulting fees from Critica (payment to a nonprofit organization). H.H. has received funding support from the Division of Microbiology and Infectious Diseases, contract no. 75N93021C00012. L.C.I. has received funding from NIH/NIAID/DMID, Moderna and Pfizer (payment made to Sanofi); grant support from GSK, Merck, Sharpe & Dohme and CDC (payment made to institution); consulting fees from Moderna Scientific Advisory Board, CDC, Pediatric Emergency Medicine Associates, American Academy of Pediatrics, Rockefeller University and American Academy of Pediatrics–Georgia Chapter; and travel support from American Academy of Pediatrics and Moderna. L.C.I. serves as in a data safety monitoring role for NIH-Phase 2 Vaccine Trial for Monkeypox, Moderna Scientific Advisory Board North America, COVID-19 Task Force, Georgia, Pediatric Infectious Disease Society, Emory University–Pediatric and Reproductive Environmental Health Scholars–Southeastern (board member), Center for Spatial Analytics, Georgia Institute of Technology (board member) and American Academy of Pediatrics (executive board for section on infectious diseases). L.A.J. has received funding from Pfizer to support a clinical trial, contract funding for research support from the CDC (to institution). Financial support for study was provided to L.A.J.’s institution by the NIH. L.A.J. also reports unpaid participation for service on data safety monitoring boards for NIH-funded clinical trials. S.K. has received funding from the NIH to conduct the COVAIL clinical trial of COVID-19 vaccines, Moderna and Janssen for COVID-19 vaccines, Pfizer to conduct clinical trials of Pfizer–BioNTech COVID-19 vaccines, and CDC to conduct surveillance for COVID-19 VE. Payments were made to Emory University. B.G.L. has received gifts from Gilead Dinner at CROI 2023. S.J.L. has received funding from NIH grants, payment made to institution. A.F.L. has received funding from Merck, Gilead and Viiv (payment to UCSF); consulting fees from Vir Biotechnology; travel support from Merck to attend a required investigator meeting; and testing kits and supplies to support research study from Hologic. K.E.L. has received funding from Pfizer–BioNTech for trial operations. M. Matkowski has received funding from the Division of Microbiology and Infectious Diseases, contract no. 75N93021C00012. T.C.S.M. has received funding from Gilead Sciences as coinvestigator and travel support for conference attendance from Gilead. M.A.M. has received funding from NIAID UM1AI148684. D.C.M. has received funding from NIH 75N93019C00050-21A: CIVICS A- Option 21A-DMID Trials of COVID-19 Vaccines. J.M. has received funding from the Division of Microbiology and Infectious Diseases contract no. 75N93021C00012. A.N. has received funding or grant support from NIH–NIAID and CEIRR, Gates Cambridge Trust and NIH–NIAID R01. K.M.N. has received grants support from the NIH to participate in the overall organization of COVID-19 trials and for participation in vaccines trials, Center for Vaccine Development and Global Health and Pfizer to conduct clinical trials of COVID-19 vaccines without salary support. R.M.N. has received funding from Moderna and Janssen, and travel support from Moderna. Z.O. has received funding from Leidos subcontract agreement no. 22CTA-DM0006 (for role as subinvestigator; clinical trial conduct, manuscript review). Payments were made to Saint Louis University. C.M.P. has received funding from NIAID UM1AI148684. R.M.P. has received funding from NIH, DMID, COVAIL, Janssen and Moderna. N.G.R. has received research grants from Pfizer, Merck, Sanofi, Quidel and Lilly. Her institution has also received funding from NIH to conduct clinical trials. N.G.R. serves on safety committees for ICON and EMMES and is on the advisory boards of Moderna and Sanofi. D.J.S. has received funding from NIH–NIAID CEIRR (NIH–NIAID R01). N.G.R. reports travel-related meetings from NIH–NIAID CEIRR. D.S.S. has received a funding grant from the NIH IDCRC. M.J.S. has received grant support from Pfizer (through institution) as a research contract for COVID-19 vaccine and therapy clinical trials, expert consulting fees from Garau Germano Medicolegal (paid to M.J.S.). M.J.S. reports serving as chair of the Section on Epidemiology, Public Health and Evidence (volunteer position) for American Academy of Pediatrics. K.T. has received funding from the Division of Microbiology and Infectious Diseases, contract no. 75N93021C00012. S.T. has received NIH–NIAID R01, CEIRR. A.W. has received funding from NIH, Sanofi and GSK (payments made to institution), and consulting fees from Aicuris, Crozet Auritec and DxNow. A.W. reports serving in a data safety monitoring role for Merck X-Vax, Vir and Curevo, and receiving other financial interest from Merck for a vaccine clinical trial. E.E.W. has received grant funding from NIH–DMID, Pfizer and Merck (with payment made to the institution), and consulting fees from Merck (payment made to E.E.W.). E.B.W. has received funding from Leidos Biomedical Research (agreement no. 22CTA-DM0009), Pfizer, Moderna, Sequiris, Clinetic and Najit Technologies (PI for vaccine study), with payments made to institution; and honoraria as a speaker from College of Diplomates of the American Board of Pediatric Dentistry (payment made to E.B.W.). He received travel support from the American Academy of Pediatrics and consulting fees from Iliad Biotechnologies. He reports serving as an advisory board member for Vaxcyte Scientific (payment made to E.B.W.). S.R.W. has received funding from NIH, Pfizer, Sanofi Pasteur, Janssen Vaccines/Johnson & Johnson, Moderna Tx, Vir Biotechnology and Worcester HIV Vaccine; and travel support from Sanofi Pasteur, with payments made to institution. S.R.W. serves in a data safety monitoring role and on an advisory board for Janssen Vaccines/Johnson & Johnson. S.R.W.’s spouse is an employee of Regeneron Pharmaceuticals and holds stock/stock options. P.L.W. has received subcontract funding from NIH, grant funding from NIH, contract funding from Pfizer to University of Iowa. P.L.W. serves in a data safety monitoring role and is an advisory board member for Emmes Corporation. The other authors declare no competing interests.
Figures







Update of
-
Bivalent and Monovalent SARS-CoV-2 Variant Vaccine Boosters Improve coverage of the known Antigenic Landscape: Results of the COVID-19 Variant Immunologic Landscape (COVAIL) Trial.Res Sq [Preprint]. 2023 May 5:rs.3.rs-2653179. doi: 10.21203/rs.3.rs-2653179/v1. Res Sq. 2023. Update in: Nat Med. 2023 Sep;29(9):2334-2346. doi: 10.1038/s41591-023-02503-4. PMID: 37205592 Free PMC article. Updated. Preprint.
References
-
- Weekly operational update on COVID-19. World Health Organizationhttps://www.who.int/emergencies/diseases/novel-coronavirus-2019/situatio... (2020).
-
- COVID-19 dashboard by the Center for Systems Science and Engineering (CCSE) at Johns Hopkins University (JHU). Johns Hopkins Universityhttps://coronavirus.jhu.edu/map.html (2022).

