Tumor-specific cholinergic CD4+ T lymphocytes guide immunosurveillance of hepatocellular carcinoma
- PMID: 37640929
- PMCID: PMC10597839
- DOI: 10.1038/s43018-023-00624-w
Tumor-specific cholinergic CD4+ T lymphocytes guide immunosurveillance of hepatocellular carcinoma
Abstract
Cholinergic nerves are involved in tumor progression and dissemination. In contrast to other visceral tissues, cholinergic innervation in the hepatic parenchyma is poorly detected. It remains unclear whether there is any form of cholinergic regulation of liver cancer. Here, we show that cholinergic T cells curtail the development of liver cancer by supporting antitumor immune responses. In a mouse multihit model of hepatocellular carcinoma (HCC), we observed activation of the adaptive immune response and induction of two populations of CD4+ T cells expressing choline acetyltransferase (ChAT), including regulatory T cells and dysfunctional PD-1+ T cells. Tumor antigens drove the clonal expansion of these cholinergic T cells in HCC. Genetic ablation of Chat in T cells led to an increased prevalence of preneoplastic cells and exacerbated liver cancer due to compromised antitumor immunity. Mechanistically, the cholinergic activity intrinsic in T cells constrained Ca2+-NFAT signaling induced by T cell antigen receptor engagement. Without this cholinergic modulation, hyperactivated CD25+ T regulatory cells and dysregulated PD-1+ T cells impaired HCC immunosurveillance. Our results unveil a previously unappreciated role for cholinergic T cells in liver cancer immunobiology.
© 2023. The Author(s).
Conflict of interest statement
T.M. owns equity in Treadwell Therapeutics, Inc., and Agios Pharmaceuticals and is a consultant for AstraZeneca. All other authors have no competing interests.
Figures
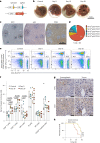
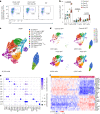


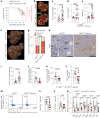


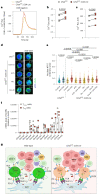









References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

