A first-in-class pan-lysyl oxidase inhibitor impairs stromal remodeling and enhances gemcitabine response and survival in pancreatic cancer
- PMID: 37640930
- PMCID: PMC10518255
- DOI: 10.1038/s43018-023-00614-y
A first-in-class pan-lysyl oxidase inhibitor impairs stromal remodeling and enhances gemcitabine response and survival in pancreatic cancer
Abstract
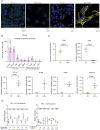
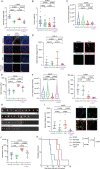
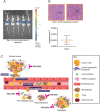
The lysyl oxidase family represents a promising target in stromal targeting of solid tumors due to the importance of this family in crosslinking and stabilizing fibrillar collagens and its known role in tumor desmoplasia. Using small-molecule drug-design approaches, we generated and validated PXS-5505, a first-in-class highly selective and potent pan-lysyl oxidase inhibitor. We demonstrate in vitro and in vivo that pan-lysyl oxidase inhibition decreases chemotherapy-induced pancreatic tumor desmoplasia and stiffness, reduces cancer cell invasion and metastasis, improves tumor perfusion and enhances the efficacy of chemotherapy in the autochthonous genetically engineered KPC model, while also demonstrating antifibrotic effects in human patient-derived xenograft models of pancreatic cancer. PXS-5505 is orally bioavailable, safe and effective at inhibiting lysyl oxidase activity in tissues. Our findings present the rationale for progression of a pan-lysyl oxidase inhibitor aimed at eliciting a reduction in stromal matrix to potentiate chemotherapy in pancreatic ductal adenocarcinoma.
© 2023. The Author(s).
Conflict of interest statement
L.P., A.D.F., B.C. and W.J. are employees of Pharmaxis, a company with ownership of PXS-5505 described in the paper. Pharmaxis provided PXS-5505 free of charge for the work presented herein. The remaining authors declare no competing interests.
Figures









References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

