Plasmodium falciparum resistant to artemisinin and diagnostics have emerged in Ethiopia
- PMID: 37640962
- PMCID: PMC10522486
- DOI: 10.1038/s41564-023-01461-4
Plasmodium falciparum resistant to artemisinin and diagnostics have emerged in Ethiopia
Abstract
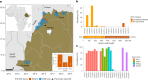
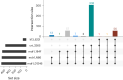
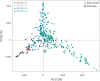
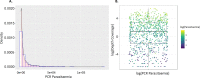
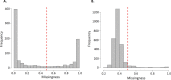
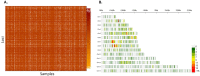
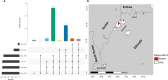
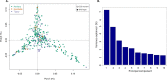
Diagnosis and treatment of Plasmodium falciparum infections are required for effective malaria control and are pre-requisites for malaria elimination efforts; hence we need to monitor emergence, evolution and spread of drug- and diagnostics-resistant parasites. We deep sequenced key drug-resistance mutations and 1,832 SNPs in the parasite genomes of 609 malaria cases collected during a diagnostic-resistance surveillance study in Ethiopia. We found that 8.0% (95% CI 7.0-9.0) of malaria cases were caused by P. falciparum carrying the candidate artemisinin partial-resistance kelch13 (K13) 622I mutation, which was less common in diagnostic-resistant parasites mediated by histidine-rich proteins 2 and 3 (pfhrp2/3) deletions than in wild-type parasites (P = 0.03). Identity-by-descent analyses showed that K13 622I parasites were significantly more related to each other than to wild type (P < 0.001), consistent with recent expansion and spread of this mutation. Pfhrp2/3-deleted parasites were also highly related, with evidence of clonal transmissions at the district level. Of concern, 8.2% of K13 622I parasites also carried the pfhrp2/3 deletions. Close monitoring of the spread of combined drug- and diagnostic-resistant parasites is needed.
© 2023. The Author(s).
Conflict of interest statement
J.B.P. reports research support from Gilead Sciences, non-financial support from Abbott Diagnostics and consulting from Zymeron Corporation, all outside the scope of the current work. All other authors have no competing interests.
Figures






References
-
- World Malaria Report 2022 (World Health Organization, 2022).
-
- Ringwald, P., Shallcross, L., Miller, J. M. & Seiber, E. Susceptibility of Plasmodium falciparum to Antimalarial Drugs: Report On Global Monitoring 1996–2004https://apps.who.int/iris/handle/10665/43302 (WHO, 2005).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

