Parkinson's disease kinase LRRK2 coordinates a cell-intrinsic itaconate-dependent defence pathway against intracellular Salmonella
- PMID: 37640963
- PMCID: PMC10962312
- DOI: 10.1038/s41564-023-01459-y
Parkinson's disease kinase LRRK2 coordinates a cell-intrinsic itaconate-dependent defence pathway against intracellular Salmonella
Abstract
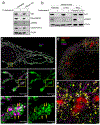
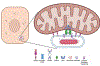
Cell-intrinsic defences constitute the first line of defence against intracellular pathogens. The guanosine triphosphatase RAB32 orchestrates one such defence response against the bacterial pathogen Salmonella, through delivery of antimicrobial itaconate. Here we show that the Parkinson's disease-associated leucine-rich repeat kinase 2 (LRRK2) orchestrates this defence response by scaffolding a complex between RAB32 and aconitate decarboxylase 1, which synthesizes itaconate from mitochondrial precursors. Itaconate delivery to Salmonella-containing vacuoles was impaired and Salmonella replication increased in LRRK2-deficient cells. Loss of LRRK2 also restored virulence of a Salmonella mutant defective in neutralizing this RAB32-dependent host defence pathway in mice. Cryo-electron tomography revealed tether formation between Salmonella-containing vacuoles and host mitochondria upon Salmonella infection, which was significantly impaired in LRRK2-deficient cells. This positions LRRK2 centrally within a host defence mechanism, which may have favoured selection of a common familial Parkinson's disease mutant allele in the human population.
© 2023. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Figures














References
-
- Tang B Rab32/38 and the xenophagic restriction of intracellular bacteria replication. Microbes Infect 18, 595–603 (2016). - PubMed
-
- Li Y et al. Analysis of the Rab GTPase Interactome in Dendritic Cells Reveals Anti-microbial Functions of the Rab32 Complex in Bacterial Containment. Immunity 44, 422–437 (2016). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

