FOXO1-regulated lncRNA CYP1B1-AS1 suppresses breast cancer cell proliferation by inhibiting neddylation
- PMID: 37640964
- PMCID: PMC10505597
- DOI: 10.1007/s10549-023-07090-z
FOXO1-regulated lncRNA CYP1B1-AS1 suppresses breast cancer cell proliferation by inhibiting neddylation
Abstract
Purpose: Overactivated neddylation is considered to be a common event in cancer. Long non-coding RNAs (lncRNAs) can regulate cancer development by mediating post-translational modifications. However, the role of lncRNA in neddylation modification remains unclear.
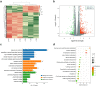
Methods: LncRNA cytochrome P450 family 1 subfamily B member 1 antisense RNA 1 (CYP1B1-AS1) expression in breast cancer tissues was evaluated by RT-PCR and TCGA BRCA data. Gain and loss of function experiments were performed to explore the role of CYP1B1-AS1 in breast cancer cell proliferation and apoptosis in vitro and in vivo. Luciferase assay, CHIP-qPCR assay, transcriptome sequencing, RNA-pulldown assay, mass spectrometry, RIP-PCR and Western blot were used to investigate the regulatory factors of CYP1B1-AS1 expression and the molecular mechanism of CYP1B1-AS1 involved in neddylation modification.
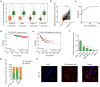
Results: We found that CYP1B1-AS1 was down-regulated in breast cancer tissues and correlated with prognosis. In vivo and in vitro functional experiments confirmed that CYP1B1-AS1 inhibited cell proliferation and induced apoptosis. Mechanistically, CYP1B1-AS1 was regulated by the transcription factor, forkhead box O1 (FOXO1), and could be upregulated by inhibiting the PI3K/FOXO1 pathway. Moreover, CYP1B1-AS1 bound directly to NEDD8 activating enzyme E1 subunit 1 (NAE1) to regulate protein neddylation.
Conclusion: This study reports for the first time that CYP1B1-AS1 inhibits protein neddylation to affect breast cancer cell proliferation, which provides a new strategy for the treatment of breast cancer by lncRNA targeting neddylation modification.
Keywords: Breast cancer; FOXO1; Long noncoding RNA; NAE1; NEDD8; Neddylation.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

