Multivariate genome-wide associations for immune traits in two maternal pig lines
- PMID: 37641029
- PMCID: PMC10463314
- DOI: 10.1186/s12864-023-09594-w
Multivariate genome-wide associations for immune traits in two maternal pig lines
Abstract
Background: Immune traits are considered to serve as potential biomarkers for pig's health. Medium to high heritabilities have been observed for some of the immune traits suggesting genetic variability of these phenotypes. Consideration of previously established genetic correlations between immune traits can be used to identify pleiotropic genetic markers. Therefore, genome-wide association study (GWAS) approaches are required to explore the joint genetic foundation for health biomarkers. Usually, GWAS explores phenotypes in a univariate (uv), trait-by-trait manner. Besides two uv GWAS methods, four multivariate (mv) GWAS approaches were applied on combinations out of 22 immune traits for Landrace (LR) and Large White (LW) pig lines.
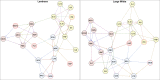
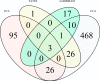
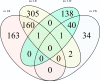
Results: In total 433 (LR: 351, LW: 82) associations were identified with the uv approach implemented in PLINK and a Bayesian linear regression uv approach (BIMBAM) software. Single Nucleotide Polymorphisms (SNPs) that were identified with both uv approaches (n = 32) were mostly associated with immune traits such as haptoglobin, red blood cell characteristics and cytokines, and were located in protein-coding genes. Mv GWAS approaches detected 647 associations for different mv immune trait combinations which were summarized to 133 Quantitative Trait Loci (QTL). SNPs for different trait combinations (n = 66) were detected with more than one mv method. Most of these SNPs are associated with red blood cell related immune trait combinations. Functional annotation of these QTL revealed 453 immune-relevant protein-coding genes. With uv methods shared markers were not observed between the breeds, whereas mv approaches were able to detect two conjoint SNPs for LR and LW. Due to unmapped positions for these markers, their functional annotation was not clarified.
Conclusions: This study evaluated the joint genetic background of immune traits in LR and LW piglets through the application of various uv and mv GWAS approaches. In comparison to uv methods, mv methodologies identified more significant associations, which might reflect the pleiotropic background of the immune system more accurately. In genetic research of complex traits, the SNP effects are generally small. Furthermore, one genetic variant can affect several correlated immune traits at the same time, termed pleiotropy. As mv GWAS methods consider strong dependencies among traits, the power to detect SNPs can be boosted. Both methods revealed immune-relevant potential candidate genes. Our results indicate that one single test is not able to detect all the different types of genetic effects in the most powerful manner and therefore, the methods should be applied complementary.
Keywords: Animal Genetics; Genome-wide Association Studies; Immune traits; Immunocompetence; Multivariate; Pigs.
© 2023. BioMed Central Ltd., part of Springer Nature.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Genome-wide associations for immune traits in two maternal pig lines.BMC Genomics. 2021 Oct 5;22(1):717. doi: 10.1186/s12864-021-07997-1. BMC Genomics. 2021. PMID: 34610786 Free PMC article.
-
Genomic background and genetic relationships between boar taint and fertility traits in German Landrace and Large White.BMC Genet. 2020 Jun 8;21(1):61. doi: 10.1186/s12863-020-00865-z. BMC Genet. 2020. PMID: 32513168 Free PMC article.
-
Comparison of two multi-trait association testing methods and sequence-based fine mapping of six additive QTL in Swiss Large White pigs.BMC Genomics. 2023 Apr 10;24(1):192. doi: 10.1186/s12864-023-09295-4. BMC Genomics. 2023. PMID: 37038103 Free PMC article.
-
Comprehensive identification of pleiotropic loci for body fat distribution using the NHGRI-EBI Catalog of published genome-wide association studies.Obes Rev. 2019 Mar;20(3):385-406. doi: 10.1111/obr.12806. Epub 2018 Nov 22. Obes Rev. 2019. PMID: 30565845 Review.
-
Underlying genetic architecture of resistance to mastitis in dairy cattle: A systematic review and gene prioritization analysis of genome-wide association studies.J Dairy Sci. 2023 Jan;106(1):323-351. doi: 10.3168/jds.2022-21923. Epub 2022 Nov 1. J Dairy Sci. 2023. PMID: 36333139
Cited by
-
Identification of SNPs and Candidate Genes Associated with Monocyte/Lymphocyte Ratio and Neutrophil/Lymphocyte Ratio in Duroc × Erhualian F2 Population.Int J Mol Sci. 2024 Sep 9;25(17):9745. doi: 10.3390/ijms25179745. Int J Mol Sci. 2024. PMID: 39273692 Free PMC article.
-
Genomic Variants Associated with Haematological Parameters and T Lymphocyte Subpopulations in a Large White and Min Pig Intercross Population.Animals (Basel). 2024 Nov 1;14(21):3140. doi: 10.3390/ani14213140. Animals (Basel). 2024. PMID: 39518863 Free PMC article.
References
-
- Heuß EM, Pröll-Cornelissen MJ, Neuhoff C, Tholen E, Große-Brinkhaus C. Invited review: piglet survival: benefits of the immunocompetence. Animal. 2019;1–11. 10.1017/S1751731119000430. - PubMed
-
- Clapperton M, Diack AB, Matika O, Glass EJ, Gladney CD, Mellencamp MA, et al. Traits associated with innate and adaptive immunity in pigs: heritability and associations with performance under different health status conditions. Genet Sel Evol. 2009;41:54. doi: 10.1186/1297-9686-41-54. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

