Emergence of the Dickeya genus involved duplication of the OmpF porin and the adaptation of the EnvZ-OmpR signaling network
- PMID: 37642428
- PMCID: PMC10581057
- DOI: 10.1128/spectrum.00833-23
Emergence of the Dickeya genus involved duplication of the OmpF porin and the adaptation of the EnvZ-OmpR signaling network
Abstract
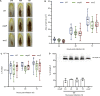
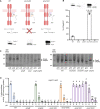
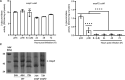
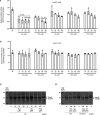
Genome evolution, and more specifically gene duplication, is a key process shaping host-microorganism interaction. The conserved paralogs usually provide an advantage to the bacterium to thrive. If not, these genes become pseudogenes and disappear. Here, we show that during the emergence of the genus Dickeya, the gene encoding the porin OmpF was duplicated. Our results show that the ompF2 expression is deleterious to the virulence of Dickeya dadantii, the agent causing soft rot disease. Interestingly, ompF2 is regulated while ompF is constitutive but activated by the EnvZ-OmpR two-component system. In vitro, acidic pH triggers the system. The pH measured in four eudicotyledons increased from an initial pH of 5.5 to 7 within 8 h post-infection. Then, the pH decreased to 5.5 at 10 h post-infection and until full maceration of the plant tissue. Yet, the production of phenolic acids by the plant's defenses prevents the activation of the EnvZ-OmpR system to avoid the ompF2 expression even though environmental conditions should trigger this system. We highlight that gene duplication in a pathogen is not automatically an advantage for the infectious process and that, there was a need for our model organism to adapt its genetic regulatory networks to conserve these duplicated genes. IMPORTANCE Dickeya species cause various diseases in a wide range of crops and ornamental plants. Understanding the molecular program that allows the bacterium to colonize the plant is key to developing new pest control methods. Unlike other enterobacterial pathogens, Dickeya dadantii, the causal agent of soft rot disease, does not require the EnvZ-OmpR system for virulence. Here, we showed that during the emergence of the genus Dickeya, the gene encoding the porin OmpF was duplicated and that the expression of ompF2 was deleterious for virulence. We revealed that while the EnvZ-OmpR system was activated in vitro by acidic pH and even though the pH was acidic when the plant is colonized, this system was repressed by phenolic acid (generated by the plant's defenses). These results provide a unique- biologically relevant-perspective on the consequence of gene duplication and the adaptive nature of regulatory networks to retain the duplicated gene.
Keywords: Dickeya dadantii; EnvZ-OmpR; phenolic acids; plant pathogen; porin; two-component system.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
LinkOut - more resources
Full Text Sources

