Isolation and genome sequencing of cytomegaloviruses from Natal multimammate mice (Mastomys natalensis)
- PMID: 37643006
- PMCID: PMC10721045
- DOI: 10.1099/jgv.0.001873
Isolation and genome sequencing of cytomegaloviruses from Natal multimammate mice (Mastomys natalensis)
Abstract
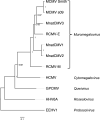
Distinct cytomegaloviruses (CMVs) are widely distributed across their mammalian hosts in a highly host species-restricted pattern. To date, evidence demonstrating this has been limited largely to PCR-based approaches targeting small, conserved genomic regions, and only a few complete genomes of isolated viruses representing distinct CMV species have been sequenced. We have now combined direct isolation of infectious viruses from tissues with complete genome sequencing to provide a view of CMV diversity in a wild animal population. We targeted Natal multimammate mice (Mastomys natalensis), which are common in sub-Saharan Africa, are known to carry a variety of zoonotic pathogens, and are regarded as the primary source of Lassa virus (LASV) spillover into humans. Using transformed epithelial cells prepared from M. natalensis kidneys, we isolated CMVs from the salivary gland tissue of 14 of 37 (36 %) animals from a field study site in Mali. Genome sequencing showed that these primary isolates represent three different M. natalensis CMVs (MnatCMVs: MnatCMV1, MnatCMV2 and MnatCMV3), with some animals carrying multiple MnatCMVs or multiple strains of a single MnatCMV presumably as a result of coinfection or superinfection. Including primary isolates and plaque-purified isolates, we sequenced and annotated the genomes of two MnatCMV1 strains (derived from sequencing 14 viruses), six MnatCMV2 strains (25 viruses) and ten MnatCMV3 strains (21 viruses), totalling 18 MnatCMV strains isolated as 60 infectious viruses. Phylogenetic analysis showed that these MnatCMVs group with other murid viruses in the genus Muromegalovirus (subfamily Betaherpesvirinae, family Orthoherpesviridae), and that MnatCMV1 and MnatCMV2 are more closely related to each other than to MnatCMV3. The availability of MnatCMV isolates and the characterization of their genomes will serve as the prelude to the generation of a MnatCMV-based vaccine to target LASV in the M. natalensis reservoir.
Keywords: Mastomys natalensis; Mastomys natalensis cytomegalovirus; cytomegalovirus; genome sequence; herpesvirus; muromegalovirus.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures



Similar articles
-
Molecular cloning and host range analysis of three cytomegaloviruses from Mastomys natalensis.J Virol. 2025 May 20;99(5):e0214724. doi: 10.1128/jvi.02147-24. Epub 2025 Apr 9. J Virol. 2025. PMID: 40202317 Free PMC article.
-
Multiple DNA viruses identified in multimammate mouse (Mastomys natalensis) populations from across regions of sub-Saharan Africa.Arch Virol. 2020 Oct;165(10):2291-2299. doi: 10.1007/s00705-020-04738-9. Epub 2020 Aug 4. Arch Virol. 2020. PMID: 32754877 Free PMC article.
-
Inoculation route-dependent Lassa virus dissemination and shedding dynamics in the natural reservoir - Mastomys natalensis.Emerg Microbes Infect. 2021 Dec;10(1):2313-2325. doi: 10.1080/22221751.2021.2008773. Emerg Microbes Infect. 2021. PMID: 34792436 Free PMC article.
-
Mapping the zoonotic niche of Lassa fever in Africa.Trans R Soc Trop Med Hyg. 2015 Aug;109(8):483-92. doi: 10.1093/trstmh/trv047. Epub 2015 Jun 17. Trans R Soc Trop Med Hyg. 2015. PMID: 26085474 Free PMC article. Review.
-
Lassa virus diversity and feasibility for universal prophylactic vaccine.F1000Res. 2019 Jan 31;8:F1000 Faculty Rev-134. doi: 10.12688/f1000research.16989.1. eCollection 2019. F1000Res. 2019. PMID: 30774934 Free PMC article. Review.
Cited by
-
Scanning the Horizon for Environmental Applications of Genetically Modified Viruses Reveals Challenges for Their Environmental Risk Assessment.Int J Mol Sci. 2024 Jan 25;25(3):1507. doi: 10.3390/ijms25031507. Int J Mol Sci. 2024. PMID: 38338787 Free PMC article. Review.
-
Molecular cloning and host range analysis of three cytomegaloviruses from Mastomys natalensis.J Virol. 2025 May 20;99(5):e0214724. doi: 10.1128/jvi.02147-24. Epub 2025 Apr 9. J Virol. 2025. PMID: 40202317 Free PMC article.
-
Fine-tuning the evolutionary stability of recombinant herpesviral transmissible vaccines.Proc Biol Sci. 2024 Nov;291(2034):20241827. doi: 10.1098/rspb.2024.1827. Epub 2024 Nov 13. Proc Biol Sci. 2024. PMID: 39532136 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

