ST6GAL1 sialyltransferase promotes acinar to ductal metaplasia and pancreatic cancer progression
- PMID: 37643018
- PMCID: PMC10619436
- DOI: 10.1172/jci.insight.161563
ST6GAL1 sialyltransferase promotes acinar to ductal metaplasia and pancreatic cancer progression
Abstract
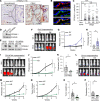
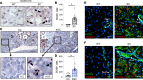
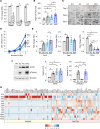
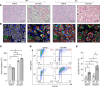
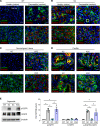
The role of aberrant glycosylation in pancreatic ductal adenocarcinoma (PDAC) remains an under-investigated area of research. In this study, we determined that ST6 β-galactoside α2,6 sialyltransferase 1 (ST6GAL1), which adds α2,6-linked sialic acids to N-glycosylated proteins, was upregulated in patients with early-stage PDAC and was further increased in advanced disease. A tumor-promoting function for ST6GAL1 was elucidated using tumor xenograft experiments with human PDAC cells. Additionally, we developed a genetically engineered mouse (GEM) model with transgenic expression of ST6GAL1 in the pancreas and found that mice with dual expression of ST6GAL1 and oncogenic KRASG12D had greatly accelerated PDAC progression compared with mice expressing KRASG12D alone. As ST6GAL1 imparts progenitor-like characteristics, we interrogated ST6GAL1's role in acinar to ductal metaplasia (ADM), a process that fosters neoplasia by reprogramming acinar cells into ductal, progenitor-like cells. We verified ST6GAL1 promotes ADM using multiple models including the 266-6 cell line, GEM-derived organoids and tissues, and an in vivo model of inflammation-induced ADM. EGFR is a key driver of ADM and is known to be activated by ST6GAL1-mediated sialylation. Importantly, EGFR activation was dramatically increased in acinar cells and organoids from mice with transgenic ST6GAL1 expression. These collective results highlight a glycosylation-dependent mechanism involved in early stages of pancreatic neoplasia.
Keywords: Cancer; Glycobiology; Oncogenes; Oncology.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

