Comparative analysis of the myoglobin gene in whales and humans reveals evolutionary changes in regulatory elements and expression levels
- PMID: 37643191
- PMCID: PMC10464968
- DOI: 10.1371/journal.pone.0284834
Comparative analysis of the myoglobin gene in whales and humans reveals evolutionary changes in regulatory elements and expression levels
Abstract
Cetacea and other diving mammals have undergone numerous adaptations to their aquatic environment, among them high levels of the oxygen-carrying intracellular hemoprotein myoglobin in skeletal muscles. Hypotheses regarding the mechanisms leading to these high myoglobin levels often invoke the induction of gene expression by exercise, hypoxia, and other physiological gene regulatory pathways. Here we explore an alternative hypothesis: that cetacean myoglobin genes have evolved high levels of transcription driven by the intrinsic developmental mechanisms that drive muscle cell differentiation. We have used luciferase assays in differentiated C2C12 cells to test this hypothesis. Contrary to our hypothesis, we find that the myoglobin gene from the minke whale, Balaenoptera acutorostrata, shows a low level of expression, only about 8% that of humans. This low expression level is broadly shared among cetaceans and artiodactylans. Previous work on regulation of the human gene has identified a core muscle-specific enhancer comprised of two regions, the "AT element" and a C-rich sequence 5' of the AT element termed the "CCAC-box". Analysis of the minke whale gene supports the importance of the AT element, but the minke whale CCAC-box ortholog has little effect. Instead, critical positive input has been identified in a G-rich region 3' of the AT element. Also, a conserved E-box in exon 1 positively affects expression, despite having been assigned a repressive role in the human gene. Last, a novel region 5' of the core enhancer has been identified, which we hypothesize may function as a boundary element. These results illustrate regulatory flexibility during evolution. We discuss the possibility that low transcription levels are actually beneficial, and that evolution of the myoglobin protein toward enhanced stability is a critical factor in the accumulation of high myoglobin levels in adult cetacean muscle tissue.
Copyright: © 2023 Sackerson et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

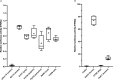


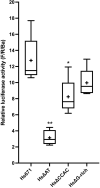


References
-
- Thewissen JGM, Cooper LN, George JC, Bajpai S. From land to water: the origin of whales, dolphins, and porpoises. Evol Edu Outreach. 2009;2:272–288. doi: 10.1007/s12052-009-0135-2 - DOI
-
- Uhen MD. The origin (s) of whales. Annu Rev Earth Planet Sci. 2010;38:189–219. doi: 10.1146/annurev-earth-040809-152453 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

