Quantitation of Circulating Mycobacterium tuberculosis Antigens by Nanopore Biosensing in Children Evaluated for Pulmonary Tuberculosis in South Africa
- PMID: 37643288
- PMCID: PMC10668583
- DOI: 10.1021/acsnano.3c04420
Quantitation of Circulating Mycobacterium tuberculosis Antigens by Nanopore Biosensing in Children Evaluated for Pulmonary Tuberculosis in South Africa
Abstract
Nanopore sensing of proteomic biomarkers lacks accuracy due to the ultralow abundance of targets, a wide variety of interferents in clinical samples, and the mismatch between pore and analyte sizes. By converting antigens to DNA probes via click chemistry and quantifying their characteristic signals, we show a nanopore assay with several amplification mechanisms to achieve an attomolar level limit of detection that enables quantitation of the circulating Mycobacterium tuberculosis (Mtb) antigen ESAT-6/CFP-10 complex in human serum. The assay's nonsputum-based feature and low-volume sample requirements make it particularly well-suited for detecting pediatric tuberculosis (TB) disease, where establishing an accurate diagnosis is greatly complicated by the paucibacillary nature of respiratory secretions, nonspecific symptoms, and challenges with sample collection. In the clinical assessment, the assay was applied to analyze ESAT-6/CFP-10 levels in serum samples collected during baseline investigation for TB in 75 children, aged 0-12 years, enrolled in a diagnostic study conducted in Cape Town, South Africa. This nanopore assay showed superior sensitivity in children with confirmed TB (94.4%) compared to clinical "gold standard" diagnostic technologies (Xpert MTB/RIF 44.4% and Mtb culture 72.2%) and filled the diagnostic gap for children with unconfirmed TB, where these traditional technologies fell short. We envision that, in combination with automated sample processing and portable nanopore devices, this methodology will offer a powerful tool to support the diagnosis of pulmonary TB in children.
Keywords: CFP-10; ESAT-6; blood test; nanopore; pediatric tuberculosis.
Figures
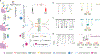
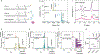


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

