Targeting Aggressive B-cell Lymphomas through Pharmacological Activation of the Mitochondrial Protease OMA1
- PMID: 37643767
- PMCID: PMC10723637
- DOI: 10.1158/1535-7163.MCT-22-0718
Targeting Aggressive B-cell Lymphomas through Pharmacological Activation of the Mitochondrial Protease OMA1
Abstract
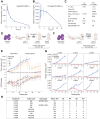
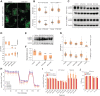
DLBCL are aggressive, rapidly proliferating tumors that critically depend on the ATF4-mediated integrated stress response (ISR) to adapt to stress caused by uncontrolled growth, such as hypoxia, amino acid deprivation, and accumulation of misfolded proteins. Here, we show that ISR hyperactivation is a targetable liability in DLBCL. We describe a novel class of compounds represented by BTM-3528 and BTM-3566, which activate the ISR through the mitochondrial protease OMA1. Treatment of tumor cells with compound leads to OMA1-dependent cleavage of DELE1 and OPA1, mitochondrial fragmentation, activation of the eIF2α-kinase HRI, cell growth arrest, and apoptosis. Activation of OMA1 by BTM-3528 and BTM-3566 is mechanistically distinct from inhibitors of mitochondrial electron transport, as the compounds induce OMA1 activity in the absence of acute changes in respiration. We further identify the mitochondrial protein FAM210B as a negative regulator of BTM-3528 and BTM-3566 activity. Overexpression of FAM210B prevents both OMA1 activation and apoptosis. Notably, FAM210B expression is nearly absent in healthy germinal center B-lymphocytes and in derived B-cell malignancies, revealing a fundamental molecular vulnerability which is targeted by BTM compounds. Both compounds induce rapid apoptosis across diverse DLBCL lines derived from activated B-cell, germinal center B-cell, and MYC-rearranged lymphomas. Once-daily oral dosing of BTM-3566 resulted in complete regression of xenografted human DLBCL SU-DHL-10 cells and complete regression in 6 of 9 DLBCL patient-derived xenografts. BTM-3566 represents a first-of-its kind approach of selectively hyperactivating the mitochondrial ISR for treating DLBCL.
©2023 American Association for Cancer Research.
Figures




References
-
- Martínez-Reyes I, Chandel NS. Cancer metabolism: looking forward. Nat Rev Cancer 2021;21:669–80. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

