Diagnostic implications of pitfalls in causal variant identification based on 4577 molecularly characterized families
- PMID: 37644014
- PMCID: PMC10465531
- DOI: 10.1038/s41467-023-40909-3
Diagnostic implications of pitfalls in causal variant identification based on 4577 molecularly characterized families
Abstract
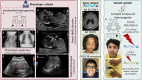
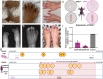
Despite large sequencing and data sharing efforts, previously characterized pathogenic variants only account for a fraction of Mendelian disease patients, which highlights the need for accurate identification and interpretation of novel variants. In a large Mendelian cohort of 4577 molecularly characterized families, numerous scenarios in which variant identification and interpretation can be challenging are encountered. We describe categories of challenges that cover the phenotype (e.g. novel allelic disorders), pedigree structure (e.g. imprinting disorders masquerading as autosomal recessive phenotypes), positional mapping (e.g. double recombination events abrogating candidate autozygous intervals), gene (e.g. novel gene-disease assertion) and variant (e.g. complex compound inheritance). Overall, we estimate a probability of 34.3% for encountering at least one of these challenges. Importantly, our data show that by only addressing non-sequencing-based challenges, around 71% increase in the diagnostic yield can be expected. Indeed, by applying these lessons to a cohort of 314 cases with negative clinical exome or genome reports, we could identify the likely causal variant in 54.5%. Our work highlights the need to have a thorough approach to undiagnosed diseases by considering a wide range of challenges rather than a narrow focus on sequencing technologies. It is hoped that by sharing this experience, the yield of undiagnosed disease programs globally can be improved.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Shickh S, Mighton C, Uleryk E, Pechlivanoglou P, Bombard Y. The clinical utility of exome and genome sequencing across clinical indications: a systematic review. Hum. Genet. 2021;140:1403–1416. - PubMed
-
- Bodian DL, Kothiyal P, Hauser NS. Pitfalls of clinical exome and gene panel testing: alternative transcripts. Genet. Med. 2019;21:1240–1245. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

