Importance of residue 248 in Escherichia coli RNase P RNA mediated cleavage
- PMID: 37644068
- PMCID: PMC10465520
- DOI: 10.1038/s41598-023-41203-4
Importance of residue 248 in Escherichia coli RNase P RNA mediated cleavage
Abstract
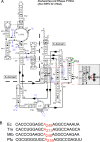
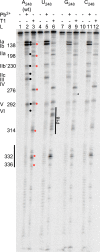
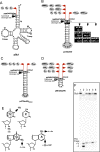
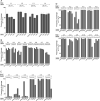
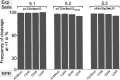
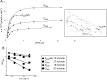
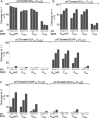
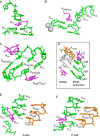
tRNA genes are transcribed as precursors and RNase P generates the matured 5' end of tRNAs. It has been suggested that residue - 1 (the residue immediately 5' of the scissile bond) in the pre-tRNA interacts with the well-conserved bacterial RNase P RNA (RPR) residue A248 (Escherichia coli numbering). The way A248 interacts with residue - 1 is not clear. To gain insight into the role of A248, we analyzed cleavage as a function of A248 substitutions and N-1 nucleobase identity by using pre-tRNA and three model substrates. Our findings are consistent with a model where the structural topology of the active site varies and depends on the identity of the nucleobases at, and in proximity to, the cleavage site and their potential to interact. This leads to positioning of Mg2+ that activates the water that acts as the nucleophile resulting in efficient and correct cleavage. We propose that in addition to be involved in anchoring the substrate the role of A248 is to exclude bulk water from access to the amino acid acceptor stem, thereby preventing non-specific hydrolysis of the pre-tRNA. Finally, base stacking is discussed as a way to protect functionally important base-pairing interactions from non-specific hydrolysis, thereby ensuring high fidelity during RNA processing and the decoding of mRNA.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Liu F, Altman SR. Protein Reviews. Springer; 2010.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

