Evaluation of enterotoxigenic Bacteroides fragilis correlation with the expression of cellular signaling pathway genes in Iranian patients with colorectal cancer
- PMID: 37644520
- PMCID: PMC10463534
- DOI: 10.1186/s13027-023-00523-w
Evaluation of enterotoxigenic Bacteroides fragilis correlation with the expression of cellular signaling pathway genes in Iranian patients with colorectal cancer
Abstract
Background: Colorectal cancer (CRC) is one of the most common cancers all over the world, and dysbiosis in the gut microbiota may play a role in colorectal carcinogenesis. Bacteroides fragilis can lead to tumorigenesis by changing signaling pathways, including the WNT/β-catenin pathway. Therefore, in the present study, we investigated the correlation between the enterotoxigenic B. fragilis amount and the expression of signaling pathway genes involved in CRC.
Materials and methods: B. fragilis was determined in 30 tumors and adjacent healthy tissues by the qPCR method. Next, the relationship between enterotoxigenic B. fragilis and the expression of signaling pathway genes, including CCND1, TP53, BCL2, BAX, WNT, TCF, AXIN, APC, and CTNNB1 was investigated. Additionally, possible correlations between clinicopathological features of the tumor samples and the abundance of B. fragilis were analyzed.
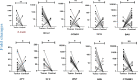
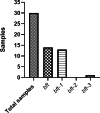
Results: The results showed that B. fragilis was detected in 100% of tumor samples and 86% of healthy tissues. Additionally, enterotoxigenic B. fragilis colonized 47% of all samples, and bft-1 toxin was the most frequently found isotype among the samples. The analysis showed that the high level of B. fragilis has a significant relationship with the high expression of AXIN, CTNNB1, and BCL2 genes. On the other hand, our results did not show any possible correlation between this bacterium and the clinicopathological features of the tumor sample.
Conclusion: B. fragilis had a higher abundance in the tumor samples than in healthy tissues, and this bacterium may lead to CRC by making changes in cellular signaling pathways and genes. Therefore, to better understand the physiological effects of B. fragilis on the inflammatory response and CRC, future research should focus on dissecting the molecular mechanisms by which this bacterium regulates cellular signaling pathways.
Keywords: BCL2; Bacteroides fragilis; Colorectal cancer; TP53; WNT/β-catenin.
© 2023. BioMed Central Ltd., part of Springer Nature.
Conflict of interest statement
The authors declare that there are no potential conflicts of interest in the present study.
The authors declare no competing interests.
Figures
Similar articles
-
Association between colorectal cancer and Fusobacterium nucleatum and Bacteroides fragilis bacteria in Iranian patients: a preliminary study.Infect Agent Cancer. 2021 Jun 9;16(1):41. doi: 10.1186/s13027-021-00381-4. Infect Agent Cancer. 2021. PMID: 34108031 Free PMC article.
-
The association between fecal enterotoxigenic B. fragilis with colorectal cancer.BMC Cancer. 2019 Sep 5;19(1):879. doi: 10.1186/s12885-019-6115-1. BMC Cancer. 2019. PMID: 31488085 Free PMC article.
-
Comparison of microbiological profile of enterotoxigenic Bacteroides fragilis (ETBF) isolates from subjects with colorectal cancer (CRC) or intestinal pre-cancerous lesions versus healthy individuals and evaluation of environmental factors involved in intestinal dysbiosis.Anaerobe. 2023 Aug;82:102757. doi: 10.1016/j.anaerobe.2023.102757. Epub 2023 Jun 26. Anaerobe. 2023. PMID: 37380012
-
The Mechanism of Bacteroides fragilis Toxin Contributes to Colon Cancer Formation.Malays J Med Sci. 2020 Jul;27(4):9-21. doi: 10.21315/mjms2020.27.4.2. Epub 2020 Aug 19. Malays J Med Sci. 2020. PMID: 32863742 Free PMC article. Review.
-
Good Gone Bad: One Toxin Away From Disease for Bacteroides fragilis.J Mol Biol. 2020 Feb 14;432(4):765-785. doi: 10.1016/j.jmb.2019.12.003. Epub 2019 Dec 17. J Mol Biol. 2020. PMID: 31857085 Review.
Cited by
-
Immune landscape in APC and TP53 related tumor microenvironment in colon adenocarcinoma: A bioinformatic analysis.Eur J Microbiol Immunol (Bp). 2024 Mar 12;14(2):154-165. doi: 10.1556/1886.2024.00015. Print 2024 May 14. Eur J Microbiol Immunol (Bp). 2024. PMID: 38470482 Free PMC article.
-
Exploring the interplay between Fusobacterium nucleatum with the expression of microRNA, and inflammatory mediators in colorectal cancer.Front Microbiol. 2023 Nov 23;14:1302719. doi: 10.3389/fmicb.2023.1302719. eCollection 2023. Front Microbiol. 2023. PMID: 38075864 Free PMC article.
-
Association between colorectal cancer, the frequency of Bacteroides fragilis, and the level of mismatch repair genes expression in the biopsy samples of Iranian patients.BMC Gastroenterol. 2024 Feb 23;24(1):82. doi: 10.1186/s12876-024-03169-z. BMC Gastroenterol. 2024. PMID: 38395750 Free PMC article.
-
Microbiome Dysbiosis, Dietary Intake and Lifestyle-Associated Factors Involve in Epigenetic Modulations in Colorectal Cancer: A Narrative Review.Cancer Control. 2024 Jan-Dec;31:10732748241263650. doi: 10.1177/10732748241263650. Cancer Control. 2024. PMID: 38889965 Free PMC article. Review.
-
Anaerobic gut bacteria and their potential role in the initiation, exacerbation, and development of human colorectal cancer: a narrative review.Front Cell Infect Microbiol. 2025 Jul 25;15:1559001. doi: 10.3389/fcimb.2025.1559001. eCollection 2025. Front Cell Infect Microbiol. 2025. PMID: 40786606 Free PMC article. Review.
References
-
- Abdullah M, Sukartini N, Nursyirwan SA, Pribadi RR, Maulahela H, Utari AP, et al. Gut microbiota profiles in early- and late-onset colorectal Cancer: a potential diagnostic biomarker in the future. Digestion. 2021;102(6):823–32. - PubMed
-
- Lin C, Cai X, Zhang J, Wang W, Sheng Q, Hua H, et al. Role of gut microbiota in the development and treatment of Colorectal Cancer. Digestion. 2019;100(1):72–8. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

