Evaluation of the clinical utility of extended non-invasive prenatal testing in the detection of chromosomal aneuploidy and microdeletion/microduplication
- PMID: 37644576
- PMCID: PMC10466692
- DOI: 10.1186/s40001-023-01285-2
Evaluation of the clinical utility of extended non-invasive prenatal testing in the detection of chromosomal aneuploidy and microdeletion/microduplication
Abstract
Background: With the development of whole-genome sequencing technology, non-invasive prenatal testing (NIPT) has been applied gradually to screen chromosomal microdeletions and microduplications that cannot be detected by traditional karyotyping. However, in NIPT, some false positives and false negatives occur. This study aimed to investigate the applicability of extended NIPT (NIPT-PLUS) in the detection of chromosomal aneuploidy and microdeletion/microduplication syndrome (MMS).
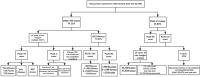
Methods: A total of 452 pregnancies that underwent prenatal diagnostic testing (amniocentesis or chorionic villus sampling) by chromosomal microarray analysis (CMA), were screened by NIPT-PLUS from the peripheral blood sample of the pregnant women. The results of the two tested items were compared and analysed.
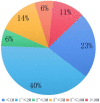
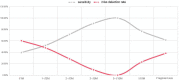
Results: Of the 452 cases, 335 (74.12%) had positive CMA results, and 117 (25.88%) had no abnormal results. A total of 86 cases of trisomy 21, 18 and 13 and sex chromosome aneuploidy (SCA) were detected by CMA and NIPT-PLUS, with a detection rate of 96.51% (83/86). Among them, the detection rates of T18, T13; 47, XXY; 47, XXX and 47 XYY were 100%, and the detection rates of T21 and 45 XO were 96.55% and 90%, respectively. The detection sensitivity of rare chromosomal trisomy (RAT) was 80% (4/5). The positive predictive values of NIPT-PLUS for chromosome aneuploidy T21, T18 and T13 and for SCA and RAT were 90.32%, 87.50%, 25.00%, 88.89% and 50%, respectively. A total of 249 cases (74.32%) of chromosomal MMS were detected by CMA. The detection rate of NIPT-PLUS was 63.86% (159/249), and 90 cases (36.14%) were missed. The larger the MMS fragment, the higher the NIPT-PLUS detection sensitivity. In addition, most small fragments were of maternal origin.
Conclusion: The comparison between the CMA and NIPT-PLUS techniques shows that NIPT-PLUS has high sensitivity for detecting chromosomal aneuploidy and chromosomal copy number variations (CNVs) with fragments > 5 M. However, the sensitivity of CNV for fragments < 5 M is low, and the missed detection rate is high. Additionally, confined placental mosaicism and foetal mosaicism are the key factors causing false negatives in NIPT-PLUS, while maternal chromosomal abnormalities and confined placental mosaicism are key contributors to false positives, so appropriate genetic counselling is especially important for pregnant women before and after NIPT-PLUS testing.
Keywords: Chromosomal microarray analysis (CMA); Microdeletion/microduplication (MMS); NIPT-PLUS; Rare chromosomal trisomy (RAT); Sex chromosome aneuploidy (SCA).
© 2023. BioMed Central Ltd., part of Springer Nature.
Conflict of interest statement
All of the authors had no any personal, financial, commercial, or academic conflicts of interest separately.
Figures
References
-
- Miller DT, Adam MP, Aradhya S, Biesecker LG, Brothman AR, Carter NP, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86(5):749–764. doi: 10.1016/j.ajhg.2010.04.006. - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources