Targeting the retinoic acid signaling pathway as a modern precision therapy against cancers
- PMID: 37645246
- PMCID: PMC10461636
- DOI: 10.3389/fcell.2023.1254612
Targeting the retinoic acid signaling pathway as a modern precision therapy against cancers
Abstract
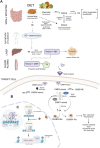
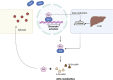
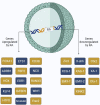
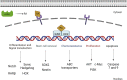
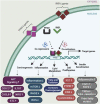
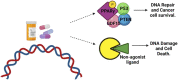
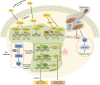
Retinoic acid (RA) is a vital metabolite derived from vitamin A. RA plays a prominent role during development, which helps in embryological advancement and cellular differentiation. Mechanistically, RA binds to its definite nuclear receptors including the retinoic acid receptor and retinoid X receptor, thus triggering gene transcription and further consequences in gene regulation. This functional heterodimer activation later results in gene activation/inactivation. Several reports have been published related to the detailed embryonic and developmental role of retinoic acids and as an anti-cancer drug for specific cancers, including acute promyelocytic leukemia, breast cancer, and prostate cancer. Nonetheless, the other side of all-trans retinoic acid (ATRA) has not been explored widely yet. In this review, we focused on the role of the RA pathway and its downstream gene activation in relation to cancer progression. Furthermore, we explored the ways of targeting the retinoic acid pathway by focusing on the dual role of aldehyde dehydrogenase (ALDH) family enzymes. Combination strategies by combining RA targets with ALDH-specific targets make the tumor cells sensitive to the treatment and improve the progression-free survival of the patients. In addition to the genomic effects of ATRA, we also highlighted the role of ATRA in non-canonical mechanisms as an immune checkpoint inhibitor, thus targeting the immune oncological perspective of cancer treatments in the current era. The role of ATRA in activating independent mechanisms is also explained in this review. This review also highlights the current clinical trials of ATRA in combination with other chemotherapeutic drugs and explains the future directional insights related to ATRA usage.
Keywords: CYP26A1; aldehyde dehydrogenases; cancer cell proliferation; chemoresistance; immune checkpoint inhibitors; retinoic acid; tumor relapse.
Copyright © 2023 Lavudi, Nuguri, Olverson, Dhanabalan, Patnaik and Kokkanti.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Regulation of a highly specific retinoic acid-4-hydroxylase (CYP26A1) enzyme and all-trans-retinoic acid metabolism in human intestinal, liver, endothelial, and acute promyelocytic leukemia cells.Leuk Lymphoma. 2005 Oct;46(10):1497-506. doi: 10.1080/10428190500174737. Leuk Lymphoma. 2005. PMID: 16194896
-
Granulocytic differentiation of human NB4 promyelocytic leukemia cells induced by all-trans retinoic acid metabolites.Cancer Res. 2001 Jan 15;61(2):700-5. Cancer Res. 2001. PMID: 11212271
-
CYP26A1 Links WNT and Retinoic Acid Signaling: A Target to Differentiate ALDH+ Stem Cells in APC-Mutant CRC.Cancers (Basel). 2024 Jan 7;16(2):264. doi: 10.3390/cancers16020264. Cancers (Basel). 2024. PMID: 38254755 Free PMC article.
-
In vitro all-trans retinoic acid (ATRA) sensitivity and cellular retinoic acid binding protein (CRABP) levels in relapse leukemic cells after remission induction by ATRA in acute promyelocytic leukemia.Leukemia. 1994;8 Suppl 2:S16-9. Leukemia. 1994. PMID: 7815831 Review.
-
In vitro all-trans retinoic acid (ATRA) sensitivity and cellular retinoic acid binding protein (CRABP) levels in relapse leukemic cells after remission induction by ATRA in acute promyelocytic leukemia.Leukemia. 1994 Jun;8(6):914-7. Leukemia. 1994. PMID: 8207983 Review.
Cited by
-
FGR Src family kinase causes signaling and phenotypic shift mimicking retinoic acid-induced differentiation of leukemic cells.Oncotarget. 2025 Mar 21;16:202-218. doi: 10.18632/oncotarget.28705. Oncotarget. 2025. PMID: 40116400 Free PMC article.
-
CD121b-positive neutrophils predict immunosuppression in septic shock.Front Immunol. 2025 Mar 31;16:1565797. doi: 10.3389/fimmu.2025.1565797. eCollection 2025. Front Immunol. 2025. PMID: 40230851 Free PMC article.
-
Electrophysiological Effects of Intratympanic Retinoic Acid Application Following Acoustic Trauma in Rats.J Int Adv Otol. 2025 May 23;21(3):1-6. doi: 10.5152/iao.2025.241834. J Int Adv Otol. 2025. PMID: 40522129 Free PMC article.
-
Combined targeting of PRDX6 and GSTP1 as a potential differentiation strategy for neuroblastoma treatment.Proc Natl Acad Sci U S A. 2025 Jun 24;122(25):e2427211122. doi: 10.1073/pnas.2427211122. Epub 2025 Jun 18. Proc Natl Acad Sci U S A. 2025. PMID: 40531876 Free PMC article.
-
Deciphering Acute Myeloid Leukemia Associated Transcription Factors in Human Primary CD34+ Hematopoietic Stem/Progenitor Cells.Cells. 2023 Dec 29;13(1):78. doi: 10.3390/cells13010078. Cells. 2023. PMID: 38201282 Free PMC article. Review.
References
Publication types
LinkOut - more resources
Full Text Sources

