Profiling neurotransmitter-evoked glial responses by RNA-sequencing analysis
- PMID: 37645568
- PMCID: PMC10461064
- DOI: 10.3389/fncir.2023.1252759
Profiling neurotransmitter-evoked glial responses by RNA-sequencing analysis
Abstract
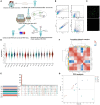
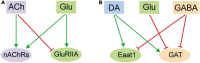
Fundamental properties of neurons and glia are distinctively different. Neurons are excitable cells that transmit information, whereas glia have long been considered as passive bystanders. Recently, the concept of tripartite synapse is proposed that glia are structurally and functionally incorporated into the synapse, the basic unit of information processing in the brains. It has then become intriguing how glia actively communicate with the presynaptic and postsynaptic compartments to influence the signal transmission. Here we present a thorough analysis at the transcriptional level on how glia respond to different types of neurotransmitters. Adult fly glia were purified from brains incubated with different types of neurotransmitters ex vivo. Subsequent RNA-sequencing analyses reveal distinct and overlapping patterns for these transcriptomes. Whereas Acetylcholine (ACh) and Glutamate (Glu) more vigorously activate glial gene expression, GABA retains its inhibitory effect. All neurotransmitters fail to trigger a significant change in the expression of their synthesis enzymes, yet Glu triggers increased expression of neurotransmitter receptors including its own and nAChRs. Expressions of transporters for GABA and Glutamate are under diverse controls from DA, GABA, and Glu, suggesting that the evoked intracellular pathways by these neurotransmitters are interconnected. Furthermore, changes in the expression of genes involved in calcium signaling also functionally predict the change in the glial activity. Finally, neurotransmitters also trigger a general metabolic suppression in glia except the DA, which upregulates a number of genes involved in transporting nutrients and amino acids. Our findings fundamentally dissect the transcriptional change in glia facing neuronal challenges; these results provide insights on how glia and neurons crosstalk in a synaptic context and underlie the mechanism of brain function and behavior.
Keywords: RNA-sequencing; glia; metabolic suppression; neurotransmitter; signal transduction.
Copyright © 2023 Wang and Ho.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Transcriptional Regulation of the Glutamate/GABA/Glutamine Cycle in Adult Glia Controls Motor Activity and Seizures in Drosophila.J Neurosci. 2019 Jul 3;39(27):5269-5283. doi: 10.1523/JNEUROSCI.1833-18.2019. Epub 2019 May 7. J Neurosci. 2019. PMID: 31064860 Free PMC article.
-
Amino acid transport in isolated neurons and glia.Adv Exp Med Biol. 1976;69:221-36. doi: 10.1007/978-1-4684-3264-0_17. Adv Exp Med Biol. 1976. PMID: 7926 Review.
-
Microglia and GABA: Diverse functions of microglia beyond GABA-receiving cells.Neurosci Res. 2023 Feb;187:52-57. doi: 10.1016/j.neures.2022.09.008. Epub 2022 Sep 21. Neurosci Res. 2023. PMID: 36152917 Review.
-
Neurotransmitters involved in fast excitatory neurotransmission directly activate enteric glial cells.Neurogastroenterol Motil. 2013 Feb;25(2):e151-60. doi: 10.1111/nmo.12065. Epub 2013 Jan 2. Neurogastroenterol Motil. 2013. PMID: 23279281
-
Calcium excitability and oscillations in suprachiasmatic nucleus neurons and glia in vitro.J Neurosci. 1992 Jul;12(7):2648-64. doi: 10.1523/JNEUROSCI.12-07-02648.1992. J Neurosci. 1992. PMID: 1351936 Free PMC article.
References
-
- Bonvento G., Bolanos J. P. (2021). Astrocyte-neuron metabolic cooperation shapes brain activity. Cell Metab. 33 1546–1564. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

