Intrinsic determinants of prion protein neurotoxicity in Drosophila: from sequence to (dys)function
- PMID: 37645703
- PMCID: PMC10461008
- DOI: 10.3389/fnmol.2023.1231079
Intrinsic determinants of prion protein neurotoxicity in Drosophila: from sequence to (dys)function
Abstract
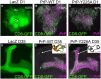
Prion diseases are fatal brain disorders characterized by deposition of insoluble isoforms of the prion protein (PrP). The normal and pathogenic structures of PrP are relatively well known after decades of studies. Yet our current understanding of the intrinsic determinants regulating PrP misfolding are largely missing. A 3D subdomain of PrP comprising the β2-α2 loop and helix 3 contains high sequence and structural variability among animals and has been proposed as a key domain regulating PrP misfolding. We combined in vivo work in Drosophila with molecular dynamics (MD) simulations, which provide additional insight to assess the impact of candidate substitutions in PrP from conformational dynamics. MD simulations revealed that in human PrP WT the β2-α2 loop explores multiple β-turn conformations, whereas the Y225A (rabbit PrP-like) substitution strongly favors a 310-turn conformation, a short right-handed helix. This shift in conformational diversity correlates with lower neurotoxicity in flies. We have identified additional conformational features and candidate amino acids regulating the high toxicity of human PrP and propose a new strategy for testing candidate modifiers first in MD simulations followed by functional experiments in flies. In this review we expand on these new results to provide additional insight into the structural and functional biology of PrP through the prism of the conformational dynamics of a 3D domain in the C-terminus. We propose that the conformational dynamics of this domain is a sensitive measure of the propensity of PrP to misfold and cause toxicity. This provides renewed opportunities to identify the intrinsic determinants of PrP misfolding through the contribution of key amino acids to different conformational states by MD simulations followed by experimental validation in transgenic flies.
Keywords: Drosophila; molecular dynamics; prion protein; protein structure; structure-function.
Copyright © 2023 Cembran and Fernandez-Funez.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


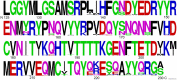



References
-
- Barlow R. M., Rennie J. C. (1976). The fate of ME7 scrapie infection in rats, guinea-pigs, and rabbits. Res. Vet. Sci. 21, 110–111. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

