This is a preprint.
Highly multiplexed mRNA quantitation with CRISPR-Cas13
- PMID: 37645785
- PMCID: PMC10461975
- DOI: 10.1101/2023.08.16.553527
Highly multiplexed mRNA quantitation with CRISPR-Cas13
Abstract
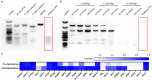
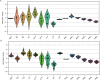
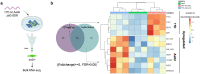
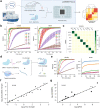
RNA quantitation tools are often either high-throughput or cost-effective, but rarely are they both. Existing methods can profile the transcriptome at great expense or are limited to quantifying a handful of genes by labor constraints. A technique that permits more throughput at a reduced cost could enable multi-gene kinetic studies, gene regulatory network analysis, and combinatorial genetic screens. Here, we introduce quantitative Combinatorial Arrayed Reactions for Multiplexed Evaluation of Nucleic acids (qCARMEN): an RNA quantitation technique which leverages the programmable RNA-targeting capabilities of CRISPR-Cas13 to address this challenge by quantifying over 4,500 gene-sample pairs in a single experiment. Using qCARMEN, we studied the response profiles of interferon-stimulated genes (ISGs) during interferon (IFN) stimulation and flavivirus infection. Additionally, we observed isoform switching kinetics during epithelial-mesenchymal transition. qCARMEN is a simple and inexpensive technique that greatly enhances the scalability of RNA quantitation for novel applications with performance similar to gold-standard methods.
Conflict of interest statement
Competing interests B.K., A.P., and C.M. have filed a provisional patent application on the use of qCARMEN. None of the other authors have competing interests pursuant to results presented here.
Figures






References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
