This is a preprint.
Triaging of α-helical proteins to the mitochondrial outer membrane by distinct chaperone machinery based on substrate topology
- PMID: 37645817
- PMCID: PMC10462106
- DOI: 10.1101/2023.08.16.553624
Triaging of α-helical proteins to the mitochondrial outer membrane by distinct chaperone machinery based on substrate topology
Update in
-
Triaging of α-helical proteins to the mitochondrial outer membrane by distinct chaperone machinery based on substrate topology.Mol Cell. 2024 Mar 21;84(6):1101-1119.e9. doi: 10.1016/j.molcel.2024.01.028. Epub 2024 Feb 29. Mol Cell. 2024. PMID: 38428433
Abstract
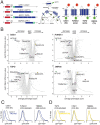
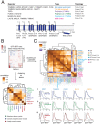
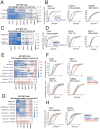
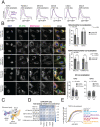
Mitochondrial outer membrane α-helical proteins play critical roles in mitochondrial-cytoplasmic communication, but the rules governing the targeting and insertion of these biophysically diverse substrates remain unknown. Here, we first defined the complement of required mammalian biogenesis machinery through genome-wide CRISPRi screens using topologically distinct membrane proteins. Systematic analysis of nine identified factors across 21 diverse α-helical substrates reveals that these components are organized into distinct targeting pathways which act on substrates based on their topology. NAC is required for efficient targeting of polytopic proteins whereas signal-anchored proteins require TTC1, a novel cytosolic chaperone which physically engages substrates. Biochemical and mutational studies reveal that TTC1 employs a conserved TPR domain and a hydrophobic groove in its C-terminal domain to support substrate solubilization and insertion into mitochondria. Thus, targeting of diverse mitochondrial membrane proteins is achieved through topological triaging in the cytosol using principles with similarities to ER membrane protein biogenesis systems.
Keywords: CRISPR; NAC; TTC1; biogenesis pathway organization; cell biology; chaperone complexes; cytosolic targeting; genetic screens; topology; α-helical outer mitochondrial membrane proteins.
Conflict of interest statement
Declaration of interests: J.S.W. declares outside interest in 5 AM Venture, Amgen, Chroma Medicine, KSQ Therapeutics, Maze Therapeutics, Tenaya Therapeutics, Tessera Therapeutics, and Third Rock Ventures. R.M.V. is a consultant and equity holder in Gate Bioscience.
Figures

References
-
- von Heijne G. (2007). The membrane protein universe: what’s out there and why bother?) J. Intern. Med. 261, 543–557. - PubMed
-
- Rapoport T.A., Li L., Park E. (2017). Structural and Mechanistic Insights into Protein Translocation. Annu. Rev. Cell. Dev. Biol. 33, 369–390. - PubMed
-
- Hegde R.S. and Keenan R.J. (2022). The mechanisms of integral membrane protein biogenesis. Nat. Rev. Mol. Cell. Biol. 23, 107–124. - PubMed
-
- Krogh A., Larsson B., von Heijne G. and Sonnhammer E. L. (2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580. - PubMed
-
- Wickner W. and Schekman R. (2005). Protein translocation across biological membranes. Science 310, 1452–1456. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
