This is a preprint.
Structure and function of the human mitochondrial MRS2 channel
- PMID: 37645897
- PMCID: PMC10462007
- DOI: 10.1101/2023.08.12.553106
Structure and function of the human mitochondrial MRS2 channel
Update in
-
Structure and function of the human mitochondrial MRS2 channel.Nat Struct Mol Biol. 2025 Mar;32(3):459-468. doi: 10.1038/s41594-024-01420-5. Epub 2024 Nov 28. Nat Struct Mol Biol. 2025. PMID: 39609651 Free PMC article.
Abstract
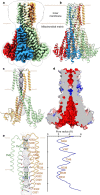
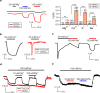
The human Mitochondrial RNA Splicing 2 protein (MRS2) has been implicated in Mg2+ transport across mitochondrial inner membranes, thus playing an important role in Mg2+ homeostasis critical for mitochondrial integrity and function. However, the molecular mechanisms underlying its fundamental channel properties such as ion selectivity and regulation remain unclear. Here, we present structural and functional investigation of MRS2. Cryo-electron microscopy structures in various ionic conditions reveal a pentameric channel architecture and the molecular basis of ion permeation and potential regulation mechanisms. Electrophysiological analyses demonstrate that MRS2 is a Ca2+-regulated, non-selective channel permeable to Mg2+, Ca2+, Na+ and K+, which contrasts with its prokaryotic ortholog, CorA, operating as a Mg2+-gated Mg2+ channel. Moreover, a conserved arginine ring within the pore of MRS2 functions to restrict cation movements, likely preventing the channel from collapsing the proton motive force that drives mitochondrial ATP synthesis. Together, our results provide a molecular framework for further understanding MRS2 in mitochondrial function and disease.
Conflict of interest statement
Competing Interests The authors declare no competing interests.
Figures
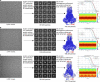
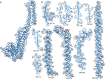
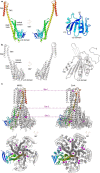

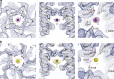



References
-
- de Baaij J. H., Hoenderop J. G. & Bindels R. J. Magnesium in man: implications for health and disease. Physiol Rev 95, 1–46 (2015). https://doi.org: 10.1152/physrev.00012.2014 - DOI - PubMed
-
- Jin F., Huang Y. & Hattori M. Recent Advances in the Structural Biology of Mg(2+) Channels and Transporters. J Mol Biol 434, 167729 (2022). https://doi.org: 10.1016/j.jmb.2022.167729 - DOI - PubMed
-
- Quamme G. A. Molecular identification of ancient and modern mammalian magnesium transporters. Am J Physiol Cell Physiol 298, C407–429 (2010). https://doi.org: 10.1152/ajpcell.00124.2009 - DOI - PubMed
-
- Wiesenberger G., Waldherr M. & Schweyen R. J. The nuclear gene MRS2 is essential for the excision of group II introns from yeast mitochondrial transcripts in vivo. J Biol Chem 267, 6963–6969 (1992). - PubMed
-
- Zsurka G., Gregan J. & Schweyen R. J. The human mitochondrial Mrs2 protein functionally substitutes for its yeast homologue, a candidate magnesium transporter. Genomics 72, 158–168 (2001). https://doi.org: 10.1006/geno.2000.6407 - DOI - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
